Pf1 filamentous bacteriophage: refinement of a molecular model by simulated annealing using 3.3 A resolution X-ray fibre diffraction data.
Gonzalez, A., Nave, C., Marvin, D.A.(1995) Acta Crystallogr D Biol Crystallogr 51: 792-804
- PubMed: 15299811
- DOI: https://doi.org/10.1107/S0907444995003027
- Primary Citation of Related Structures:
2IFM, 2IFN, 3IFM, 4IFM - PubMed Abstract:
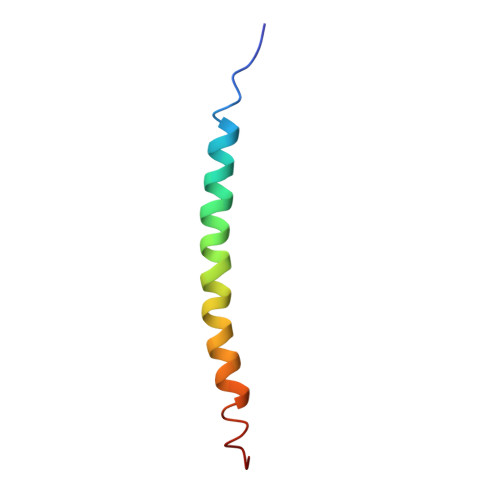
The filamentous bacteriophage Pf1 is structurally similar to the well known Ff (fd, fl, M13) strains, but it gives much better X-ray diffraction patterns, enabling a more detailed analysis of the molecular structure. The 46-residue protein subunit can be closely approximated by a single gently curved stretch of alpha-helix. The axes of the subunits are at a small angle to the virion axis, and several thousand subunits form an overlapping inter-digitated helical array surrounding a DNA core. We have derived a detailed model of the virion based on X-ray data and stereochemical constraints. We have considered potential sources of error in the diffraction data, and used the improved data to study regions where the protein subunit of Pf1 may deviate from a continuous alpha-helix. We use simulated annealing to escape from local minima, and various kinds of electron-density maps to guide the model building. Refinement of the model shows that the first few residues at the N terminus are non-helical, and there is a slight discontinuity in the alpha-helix near the middle of the sequence. The model is consistent both with general structural principles derived from high-resolution analysis of other proteins, and with specific chemical and spectroscopic data about Pf1. We apply the same refinement techniques to an alternative model with a non-helical surface loop between residues 13 and 19. Comparative analysis of models with and without a loop shows that the loop model is not supported by 3.3 A resolution X-ray diffraction data.
- Daresbury Laboratory, Warrington, England.
Organizational Affiliation: