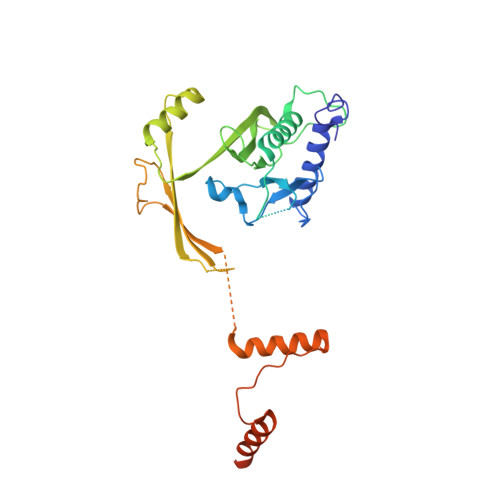
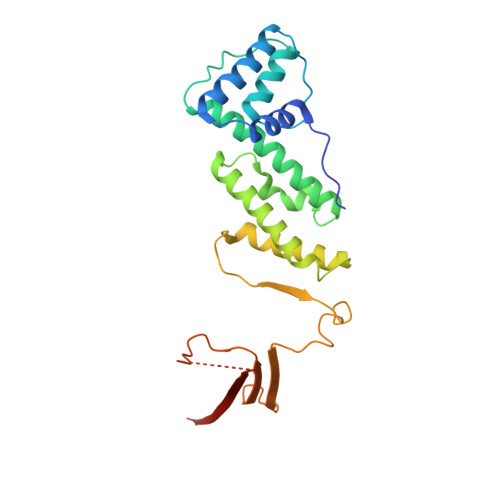
Structural analysis of the RZZ complex reveals common ancestry with multisubunit vesicle tethering machinery.
Civril, F., Wehenkel, A., Giorgi, F.M., Santaguida, S., Di Fonzo, A., Grigorean, G., Ciccarelli, F.D., Musacchio, A.(2010) Structure 18: 616-626
- PubMed: 20462495
- DOI: https://doi.org/10.1016/j.str.2010.02.014
- Primary Citation of Related Structures:
3IF8 - PubMed Abstract:
The RZZ complex recruits dynein to kinetochores. We investigated structure, topology, and interactions of the RZZ subunits (ROD, ZWILCH, and ZW10) in vitro, in vivo, and in silico. We identify neuroblastoma-amplified gene (NAG), a ZW10 binder, as a ROD homolog. ROD and NAG contain an N-terminal beta propeller followed by an alpha solenoid, which is the architecture of certain nucleoporins and vesicle coat subunits, suggesting a distant evolutionary relationship. ZW10 binding to ROD and NAG is mutually exclusive. The resulting ZW10 complexes (RZZ and NRZ) respectively contain ZWILCH and RINT1 as additional subunits. The X-ray structure of ZWILCH, the first for an RZZ subunit, reveals a novel fold distinct from RINT1's. The evolutionarily conserved NRZ likely acts as a tethering complex for retrograde trafficking of COPI vesicles from the Golgi to the endoplasmic reticulum. The RZZ, limited to metazoans, probably evolved from the NRZ, exploiting the dynein-binding capacity of ZW10 to direct dynein to kinetochores.
- Department of Experimental Oncology, European Institute of Oncology, Via Adamello 16, I-20139 Milan, Italy.
Organizational Affiliation: