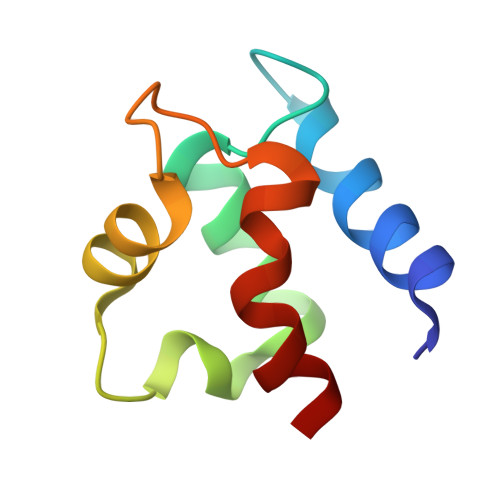
The refined structure of vitamin D-dependent calcium-binding protein from bovine intestine. Molecular details, ion binding, and implications for the structure of other calcium-binding proteins.
Szebenyi, D.M., Moffat, K.(1986) J Biological Chem 261: 8761-8777
- PubMed: 3722173
- Primary Citation of Related Structures:
3ICB - PubMed Abstract:
The structure of bovine intestinal calcium-binding protein (ICaBP) has been determined crystallographically at a resolution of 2.3 A and refined by a least squares technique to an R factor of 17.8%. The refined structure includes all 600 non-hydrogen protein atoms, two bound calcium ions, and solvent consisting of one sulfate ion and 36 water molecules. The molecule consists of two helix-loop-helix calcium-binding domains known as EF hands, connected by a linker containing a single turn of helix. Helix-helix interactions are primarily hydrophobic, but also include a few strategic hydrogen bonds. Most of the hydrogen bonds, however, are found in the calcium-binding loops, where they occur both within a single loop and between the two. Examination of the hydrogen bonding patterns in the calcium-binding loops of ICaBP and the related protein, parvalbumin, reveals several conserved hydrogen bonds which are evidently important for loop stabilization. The primary and tertiary structural features which promote the formation of an EF hand were originally identified from the structure of parvalbumin. They are modified in light of the ICaBP structure and considered as they apply to other calcium-binding proteins. The C-terminal domain of ICaBP is a normal EF hand, with ion binding properties similar to those of the calmodulin hands, but the N-terminal domain is a variant hand whose calcium ligands are mostly peptide carbonyls. Relative to a normal EF hand, this domain exhibits a similar KD for calcium binding but a greatly reduced affinity for calcium analogs such as cadmium and the lanthanide series. Lanthanides in particular may be inappropriate models for calcium in this system.