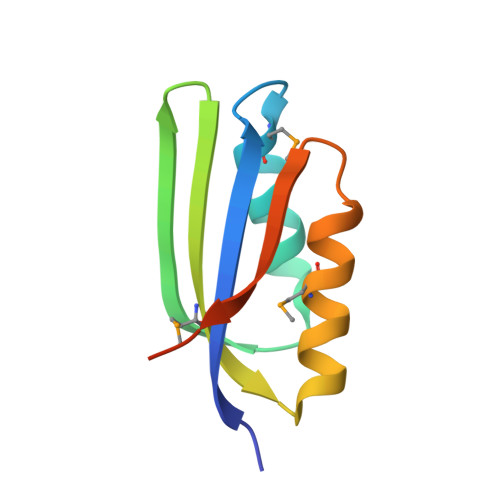
Crystal Structure of the ACT domain from GTP pyrophosphokinase of Chlorobium tepidum.
Vorobiev, S., Su, M., Seetharaman, J., Janjua, J., Xiao, R., Ciccosanti, C., Wang, H., Everett, J.K., Nair, R., Acton, T.B., Rost, B., Montelione, G.T., Tong, L., Hunt, J.F.To be published.