Novel immunodominant peptide presentation strategy: a featured HLA-A*2402-restricted cytotoxic T-lymphocyte epitope stabilized by intrachain hydrogen bonds from severe acute respiratory syndrome coronavirus nucleocapsid protein
Liu, J., Wu, P., Gao, F., Qi, J., Kawana-Tachikawa, A., Xie, J., Vavricka, C.J., Iwamoto, A., Li, T., Gao, G.F.(2010) J Virol 84: 11849-11857
- PubMed: 20844028
- DOI: https://doi.org/10.1128/JVI.01464-10
- Primary Citation of Related Structures:
3I6L - PubMed Abstract:
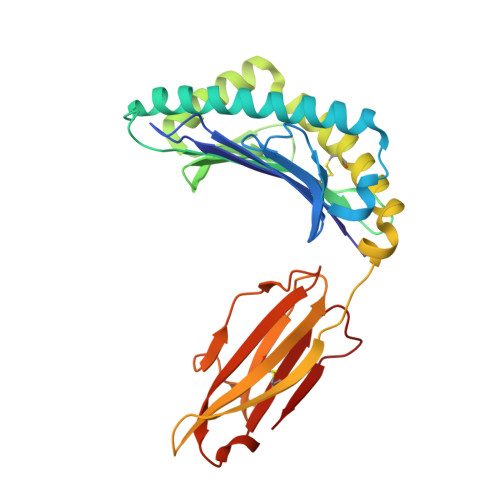
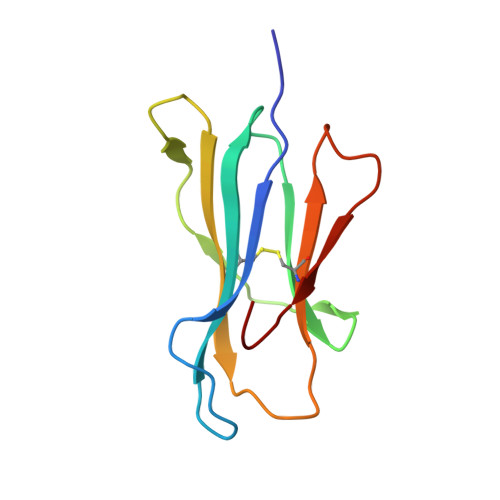
Antigenic peptides recognized by virus-specific cytotoxic T lymphocytes (CTLs) are presented by major histocompatibility complex (MHC; or human leukocyte antigen [HLA] in humans) molecules, and the peptide selection and presentation strategy of the host has been studied to guide our understanding of cellular immunity and vaccine development. Here, a severe acute respiratory syndrome coronavirus (SARS-CoV) nucleocapsid (N) protein-derived CTL epitope, N1 (QFKDNVILL), restricted by HLA-A*2402 was identified by a series of in vitro studies, including a computer-assisted algorithm for prediction, stabilization of the peptide by co-refolding with HLA-A*2402 heavy chain and β(2)-microglobulin (β(2)m), and T2-A24 cell binding. Consequently, the antigenicity of the peptide was confirmed by enzyme-linked immunospot (ELISPOT), proliferation assays, and HLA-peptide complex tetramer staining using peripheral blood mononuclear cells (PBMCs) from donors who had recovered from SARS donors. Furthermore, the crystal structure of HLA-A*2402 complexed with peptide N1 was determined, and the featured peptide was characterized with two unexpected intrachain hydrogen bonds which augment the central residues to bulge out of the binding groove. This may contribute to the T-cell receptor (TCR) interaction, showing a host immunodominant peptide presentation strategy. Meanwhile, a rapid and efficient strategy is presented for the determination of naturally presented CTL epitopes in the context of given HLA alleles of interest from long immunogenic overlapping peptides.
- Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, People's Republic of China.
Organizational Affiliation: