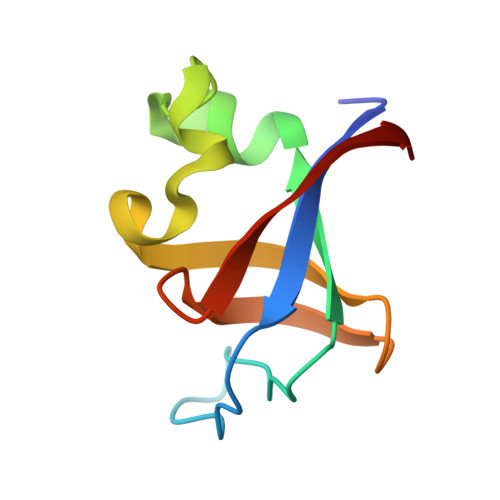
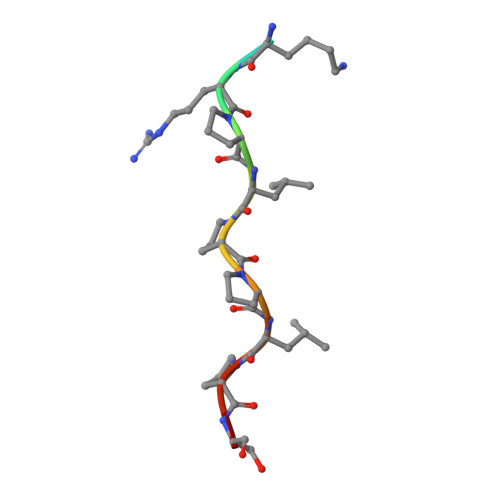
Structural studies of the phosphatidylinositol 3-kinase (PI3K) SH3 domain in complex with a peptide ligand: role of the anchor residue in ligand binding.
Batra-Safferling, R., Granzin, J., Modder, S., Hoffmann, S., Willbold, D.(2010) Biol Chem 391: 33-42
- PubMed: 19919182
- DOI: https://doi.org/10.1515/BC.2010.003
- Primary Citation of Related Structures:
3I5R, 3I5S - PubMed Abstract:
Src homology 3 (SH3) domains are mediators of protein-protein interactions. They comprise approximately 60 amino acid residues and are found in many intracellular signaling proteins. Here, we present the crystal structure of the SH3 domain from phosphatidylinositol 3-kinase (PI3K) in complex with the 12-residue proline-rich peptide PD1R (HSKRPLPPLPSL). The crystal structure of the PI3K SH3-PD1R complex at a resolution of 1.7 A reveals type I ligand orientation of the bound peptide with an extended conformation where the central portion forms a left-handed type II polyproline (PPII) helix. The overall structure of the SH3 domain shows minimal changes on ligand binding. In addition, we also attempted crystallization with another peptide ligand (PD1) where the residue at anchor position P(-3) is a tyrosine. The crystals obtained did not contain the PD1 ligand; instead, the ligand binding site is partially occupied by residues Arg18 and Trp55 from the symmetry-related PI3K SH3 molecule. Considering these crystal structures of PI3K SH3 together with published reports, we provide a comparative analysis of protein-ligand interactions that has helped us identify the individual residues which play an important role in defining target specificity.
- Institut für Strukturbiologie und Biophysik ISB-2, Forschungszentrum Jülich, D-52425 Jülich, Germany. r.batra-safferling@fz-juelich.de
Organizational Affiliation: