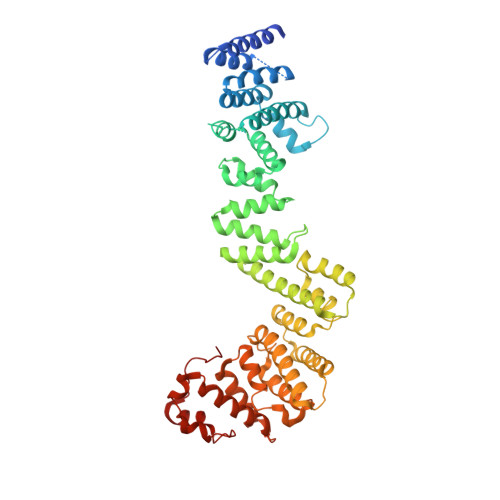
Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element.
Whittle, J.R., Schwartz, T.U.(2009) J Biological Chem 284: 28442-28452
- PubMed: 19674973
- DOI: https://doi.org/10.1074/jbc.M109.023580
- Primary Citation of Related Structures:
3I4R, 3I5P, 3I5Q - PubMed Abstract:
The nuclear pore complex (NPC) constitutes one of the largest protein assemblies in the eukaryotic cell and forms the exclusive gateway to the nucleus. The stable, approximately 15-20-MDa scaffold ring of the NPC is built from two multiprotein complexes arranged around a central 8-fold axis. Here we present crystal structures of two large architectural units, yNup170(979-1502) and hNup107(658-925) x hNup133(517-1156), each a constituent of one of the two multiprotein complexes. Conservation of domain arrangement and of tertiary structure suggests that Nup157/170 and Nup133 derived from a common ancestor. Together with the previously established ancestral coatomer element (ACE1), these two elements constitute the major alpha-helical building blocks of the NPC scaffold and define its branched, lattice-like architecture, similar to vesicle coats like COPII. We hypothesize that the extant NPC evolved early during eukaryotic evolution from a rudimentary structure composed of several identical copies of a few ancestral elements, later diversified and specified by gene duplication.
- Department of Biology, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139.
Organizational Affiliation: