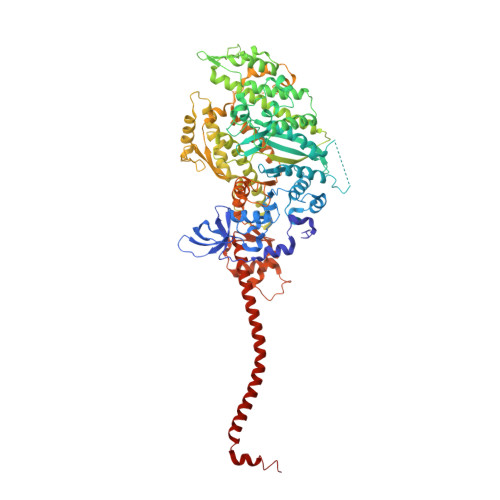
Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Yang, Y., Gourinath, S., Kovacs, M., Nyitray, L., Reutzel, R., Himmel, D.M., O'Neall-Hennessey, E., Reshetnikova, L., Szent-Gyorgyi, A.G., Brown, J.H., Cohen, C.(2007) Structure 15: 553-564
- PubMed: 17502101
- DOI: https://doi.org/10.1016/j.str.2007.03.010
- Primary Citation of Related Structures:
2EC6, 2OS8, 2OTG, 3I5F, 3I5G, 3I5H, 3I5I - PubMed Abstract:
Unlike processive cellular motors such as myosin V, whose structure has recently been determined in a "rigor-like" conformation, myosin II from contracting muscle filaments necessarily spends most of its time detached from actin. By using squid and sea scallop sources, however, we have now obtained similar rigor-like atomic structures for muscle myosin heads (S1). The significance of the hallmark closed actin-binding cleft in these crystal structures is supported here by actin/S1-binding studies. These structures reveal how different duty ratios, and hence cellular functions, of the myosin isoforms may be accounted for, in part, on the basis of detailed differences in interdomain contacts. Moreover, the rigor-like position of switch II turns out to be unique for myosin V. The overall arrangements of subdomains in the motor are relatively conserved in each of the known contractile states, and we explore qualitatively the energetics of these states.
- Rosenstiel Basic Medical Sciences Research Center, Brandeis University, Waltham, MA 02454, USA.
Organizational Affiliation: