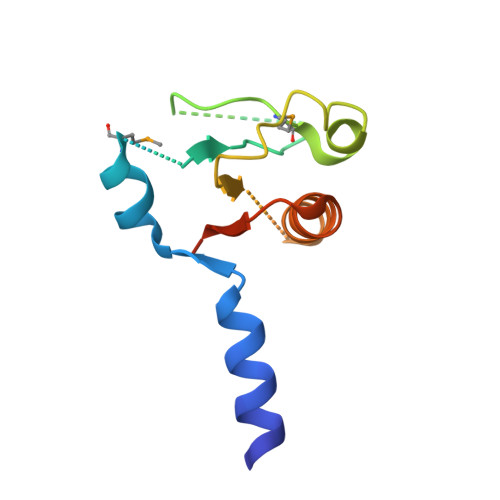
Crystal structure of lp_1913 protein from lactobacillus plantarum, northeast structural genomics Consortium target lpr140a
Seetharaman, J., Chen, Y., Forouhar, F., Sahdev, S., Janjua, H., Xiao, R., Ciccosanti, C., Foote, E.L., Acton, T.B., Rost, B., Montelione, G.T., Hunt, J.F., Tong, L.To be published.