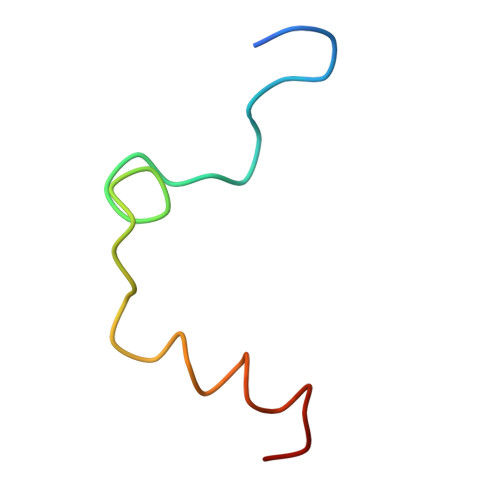
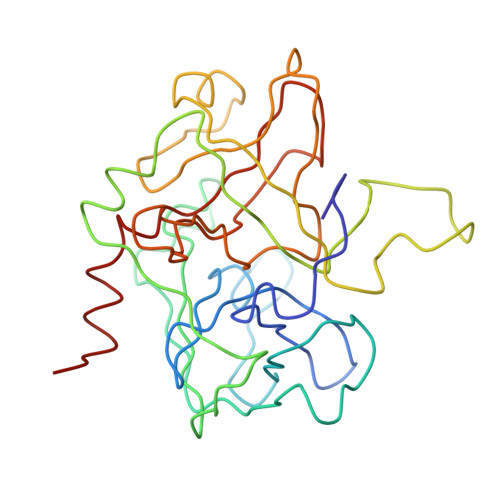
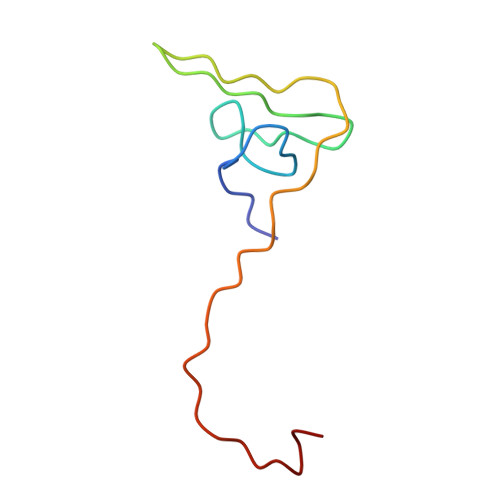
The structure of a complex of recombinant hirudin and human alpha-thrombin.
Rydel, T.J., Ravichandran, K.G., Tulinsky, A., Bode, W., Huber, R., Roitsch, C., Fenton 2nd., J.W.(1990) Science 249: 277-280
- PubMed: 2374926
- DOI: https://doi.org/10.1126/science.2374926
- Primary Citation of Related Structures:
3HTC - PubMed Abstract:
The crystallographic structure of a recombinant hirudin-thrombin complex has been solved at 2.3 angstrom (A) resolution. Hirudin consists of an NH2-terminal globular domain and a long (39 A) COOH-terminal extended domain. Residues Ile1 to Tyr3 of hirudin form a parallel beta-strand with Ser214 to Glu217 of thrombin with the nitrogen atom of Ile1 making a hydrogen bond with Ser195 O gamma atom of the catalytic site, but the specificity pocket of thrombin is not involved in the interaction. The COOH-terminal segment makes numerous electrostatic interactions with an anion-binding exosite of thrombin, whereas the last five residues are in a helical loop that forms many hydrophobic contacts. In all, 27 of the 65 residues of hirudin have contacts less than 4.0 A with thrombin (10 ion pairs and 23 hydrogen bonds). Such abundant interactions may account for the high affinity and specificity of hirudin.
- Department of Chemistry, Michigan State University, East Lansing 48824.
Organizational Affiliation: