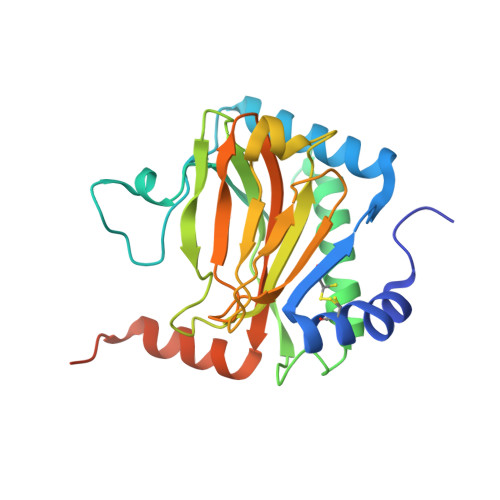
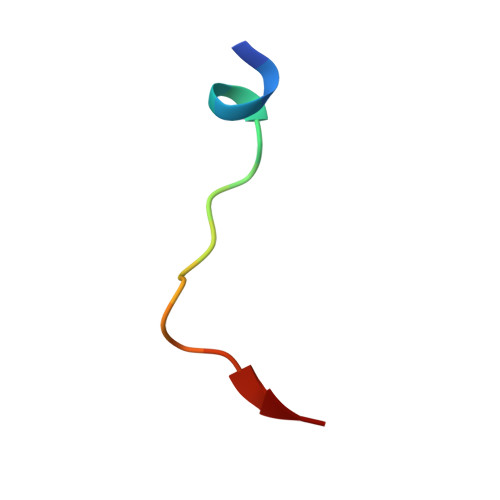
Structural basis for binding of hypoxia-inducible factor to the oxygen-sensing prolyl hydroxylases
Chowdhury, R., McDonough, M.A., Mecinovic, J., Loenarz, C., Flashman, E., Hewitson, K.S., Domene, C., Schofield, C.J.(2009) Structure 17: 981-989
- PubMed: 19604478
- DOI: https://doi.org/10.1016/j.str.2009.06.002
- Primary Citation of Related Structures:
3HQR, 3HQU - PubMed Abstract:
The oxygen-dependent hydroxylation of proline residues in the alpha subunit of hypoxia-inducible transcription factor (HIFalpha) is central to the hypoxic response in animals. Prolyl hydroxylation of HIFalpha increases its binding to the von Hippel-Lindau protein (pVHL), so signaling for degradation via the ubiquitin-proteasome system. The HIF prolyl hydroxylases (PHDs, prolyl hydroxylase domain enzymes) are related to the collagen prolyl hydroxylases, but form unusually stable complexes with their Fe(II) cofactor and 2-oxoglutarate cosubstrate. We report crystal structures of the catalytic domain of PHD2, the most important of the human PHDs, in complex with the C-terminal oxygen-dependent degradation domain of HIF-1alpha. Together with biochemical analyses, the results reveal that PHD catalysis involves a mobile region that isolates the hydroxylation site and stabilizes the PHD2.Fe(II).2OG complex. The results will be of use in the design of PHD inhibitors aimed at treating anemia and ischemic disease.
- Department of Chemistry and Oxford Centre for Integrative Systems Biology, Chemistry Research Laboratory, University of Oxford, Oxford OX1 3TA, UK.
Organizational Affiliation: