Rejuvenation of CcdB-Poisoned Gyrase by an Intrinsically Disordered Protein Domain.
De Jonge, N., Garcia-Pino, A., Buts, L., Haesaerts, S., Charlier, D., Zangger, K., Wyns, L., De Greve, H., Loris, R.(2009) Mol Cell 35: 154-163
- PubMed: 19647513
- DOI: https://doi.org/10.1016/j.molcel.2009.05.025
- Primary Citation of Related Structures:
3G7Z, 3HPW - PubMed Abstract:
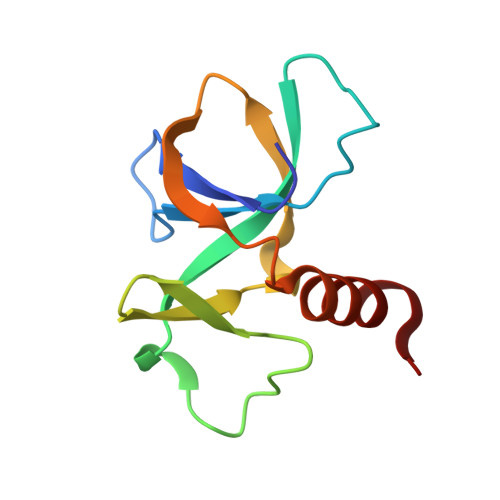
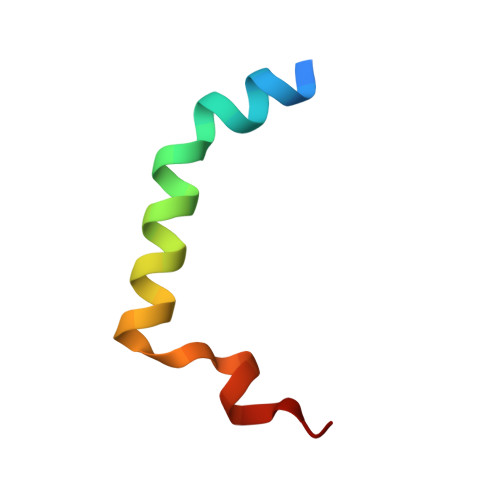
Toxin-antitoxin modules are small regulatory circuits that ensure survival of bacterial populations under challenging environmental conditions. The ccd toxin-antitoxin module on the F plasmid codes for the toxin CcdB and its antitoxin CcdA. CcdB poisons gyrase while CcdA actively dissociates CcdB:gyrase complexes in a process called rejuvenation. The CcdA:CcdB ratio modulates autorepression of the ccd operon. The mechanisms behind both rejuvenation and regulation of expression are poorly understood. We show that CcdA binds consecutively to two partially overlapping sites on CcdB, which differ in affinity by six orders of magnitude. The first, picomolar affinity interaction triggers a conformational change in CcdB that initiates the dissociation of CcdB:gyrase complexes by an allosteric segmental binding mechanism. The second, micromolar affinity binding event regulates expression of the ccd operon. Both functions of CcdA, rejuvenation and autoregulation, are mechanistically intertwined and depend crucially on the intrinsically disordered nature of the CcdA C-terminal domain.
- Structural Biology Brussels, Vrije Universiteit Brussel, Pleinlaan 2, 1050 Brussels, Belgium.
Organizational Affiliation: