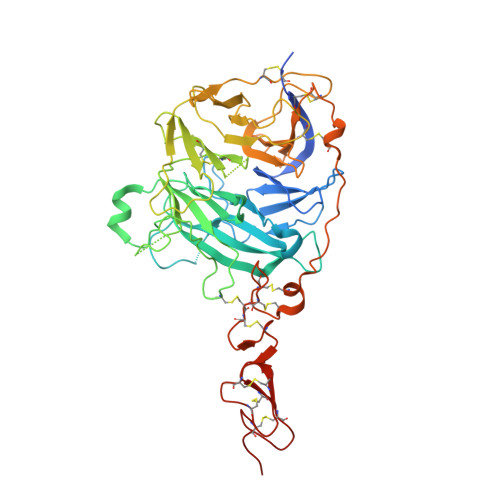
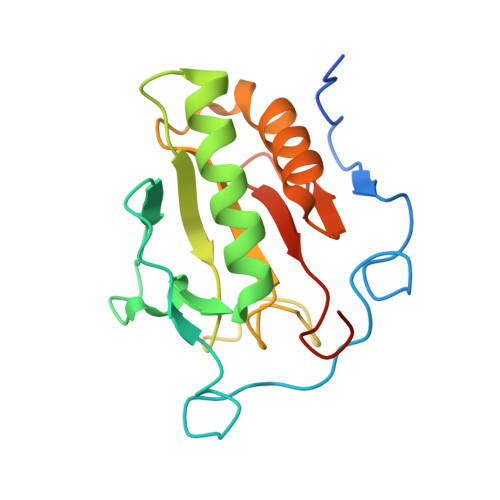
The structure of SHH in complex with HHIP reveals a recognition role for the Shh pseudo active site in signaling.
Bosanac, I., Maun, H.R., Scales, S.J., Wen, X., Lingel, A., Bazan, J.F., de Sauvage, F.J., Hymowitz, S.G., Lazarus, R.A.(2009) Nat Struct Mol Biol 16: 691-697
- PubMed: 19561609
- DOI: https://doi.org/10.1038/nsmb.1632
- Primary Citation of Related Structures:
3HO3, 3HO4, 3HO5 - PubMed Abstract:
Hedgehog (Hh) signaling is crucial for many aspects of embryonic development, whereas dysregulation of this pathway is associated with several types of cancer. Hedgehog-interacting protein (Hhip) is a surface receptor antagonist that is equipotent against all three mammalian Hh homologs. The crystal structures of human HHIP alone and bound to Sonic hedgehog (SHH) now reveal that HHIP is comprised of two EGF domains and a six-bladed beta-propeller domain. In the complex structure, a critical loop from HHIP binds the pseudo active site groove of SHH and directly coordinates its Zn2+ cation. Notably, sequence comparisons of this SHH binding loop with the Hh receptor Patched (Ptc1) ectodomains and HHIP- and PTC1-peptide binding studies suggest a 'patch for Patched' at the Shh pseudo active site; thus, we propose a role for Hhip as a structural decoy receptor for vertebrate Hh.
- Department of Structural Biology, Genentech, Inc., South San Francisco, California, USA.
Organizational Affiliation: