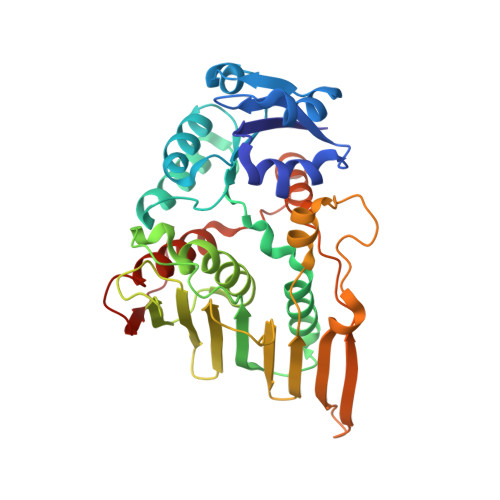
Crystal Structure of an Oxidoreductase from Bacillus cereus. Northeast Structural Genomics Consortium target id BcR251
Seetharaman, J., Su, M., Sahdev, S., Janjua, H., Xiao, R., Ciccosanti, C., Foote, E.L., Acton, T.B., Rost, B., Montelione, G.T., Tong, L., Hunt, J.F.To be published.