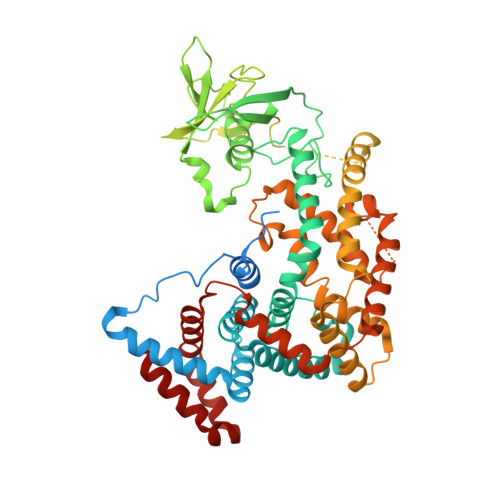
Structure and function of the intracellular region of the plexin-b1 transmembrane receptor.
Tong, Y., Hota, P.K., Penachioni, J.Y., Hamaneh, M.B., Kim, S., Alviani, R.S., Shen, L., He, H., Tempel, W., Tamagnone, L., Park, H.W., Buck, M.(2009) J Biological Chem 284: 35962-35972
- PubMed: 19843518
- DOI: https://doi.org/10.1074/jbc.M109.056275
- Primary Citation of Related Structures:
3HM6 - PubMed Abstract:
Members of the plexin family are unique transmembrane receptors in that they interact directly with Rho family small GTPases; moreover, they contain a GTPase-activating protein (GAP) domain for R-Ras, which is crucial for plexin-mediated regulation of cell motility. However, the functional role and structural basis of the interactions between the different intracellular domains of plexins remained unclear. Here we present the 2.4 A crystal structure of the complete intracellular region of human plexin-B1. The structure is monomeric and reveals that the GAP domain is folded into one structure from two segments, separated by the Rho GTPase binding domain (RBD). The RBD is not dimerized, as observed previously. Instead, binding of a conserved loop region appears to compete with dimerization and anchors the RBD to the GAP domain. Cell-based assays on mutant proteins confirm the functional importance of this coupling loop. Molecular modeling based on structural homology to p120(GAP).H-Ras suggests that Ras GTPases can bind to the plexin GAP region. Experimentally, we show that the monomeric intracellular plexin-B1 binds R-Ras but not H-Ras. These findings suggest that the monomeric form of the intracellular region is primed for GAP activity and extend a model for plexin activation.
- Departments of Physiology and Biophysics, Case Western Reserve University School of Medicine, Cleveland, Ohio 44106, USA.
Organizational Affiliation: