Structural basis of activation-dependent binding of ligand-mimetic antibody AL-57 to integrin LFA-1.
Zhang, H., Liu, J.H., Yang, W., Springer, T., Shimaoka, M., Wang, J.H.(2009) Proc Natl Acad Sci U S A 106: 18345-18350
- PubMed: 19805116
- DOI: https://doi.org/10.1073/pnas.0909301106
- Primary Citation of Related Structures:
3HI5, 3HI6 - PubMed Abstract:
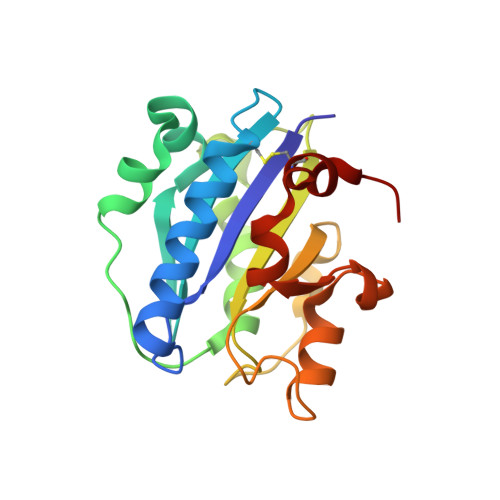
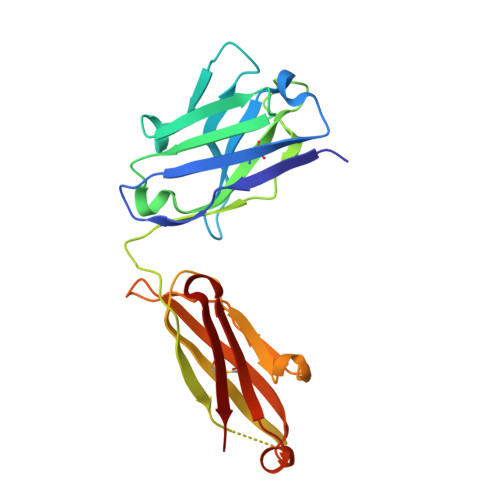
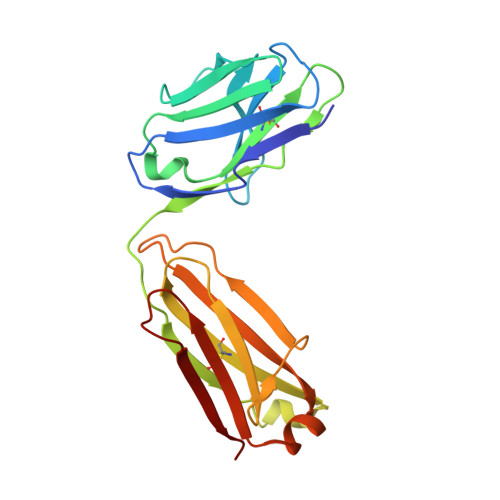
The activity of integrin LFA-1 (alpha(L)beta(2)) to its ligand ICAM-1 is regulated through the conformational changes of its ligand-binding domain, the I domain of alpha(L) chain, from an inactive, low-affinity closed form (LA), to an intermediate-affinity form (IA), and then finally, to a high-affinity open form (HA). A ligand-mimetic human monoclonal antibody AL-57 (activated LFA-1 clone 57) was identified by phage display to specifically recognize the affinity-upregulated I domain. Here, we describe the crystal structures of the Fab fragment of AL-57 in complex with IA, as well as in its unligated form. We discuss the structural features conferring AL-57's strong selectivity for the high affinity, open conformation of the I domain. The AL-57-binding site overlaps the ICAM-1 binding site on the I domain. Furthermore, an antibody Asp mimics an ICAM Glu by forming a coordination to the metal-ion dependent adhesion site (MIDAS). The structure also reveals better shape complementarity and a more hydrophobic interacting interface in AL-57 binding than in ICAM-1 binding. The results explain AL-57's antagonistic mimicry of LFA-1's natural ligands, the ICAM molecules.
- Department of Pediatrics, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA 02115, USA.
Organizational Affiliation: