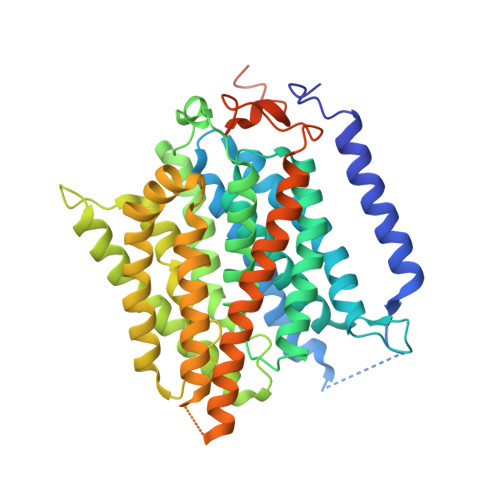
Function of human Rh based on structure of RhCG at 2.1 A.
Gruswitz, F., Chaudhary, S., Ho, J.D., Schlessinger, A., Pezeshki, B., Ho, C.M., Sali, A., Westhoff, C.M., Stroud, R.M.(2010) Proc Natl Acad Sci U S A 107: 9638-9643
- PubMed: 20457942
- DOI: https://doi.org/10.1073/pnas.1003587107
- Primary Citation of Related Structures:
3HD6 - PubMed Abstract:
In humans, NH(3) transport across cell membranes is facilitated by the Rh (rhesus) family of proteins. Human Rh C glycoprotein (RhCG) forms a trimeric complex that plays an essential role in ammonia excretion and renal pH regulation. The X-ray crystallographic structure of human RhCG, determined at 2.1 A resolution, reveals the mechanism of ammonia transport. Each monomer contains 12 transmembrane helices, one more than in the bacterial homologs. Reconstituted into proteoliposomes, RhCG conducts NH(3) to raise internal pH. Models of the erythrocyte Rh complex based on our RhCG structure suggest that the erythrocytic Rh complex is composed of stochastically assembled heterotrimers of RhAG, RhD, and RhCE.
- Department of Biochemistry and Biophysics, S412C Genentech Hall, Center for the Structure of Membrane Proteins, and Membrane Protein Expression Center, University of California, San Francisco, CA 94158, USA.
Organizational Affiliation: