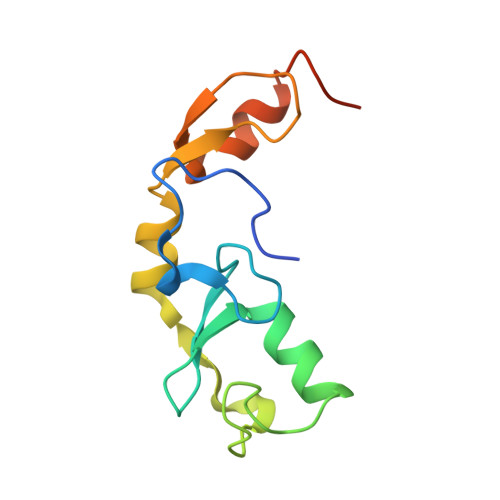
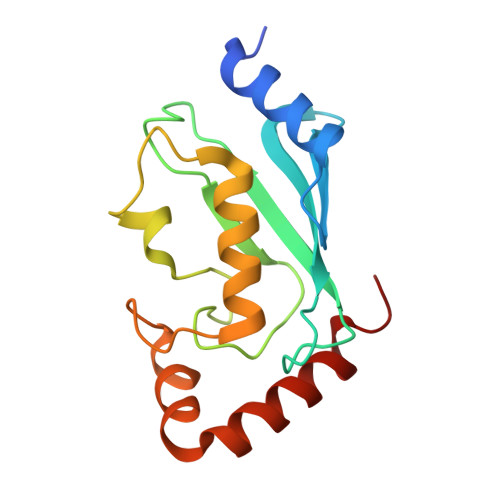
E2 interaction and dimerization in the crystal structure of TRAF6.
Yin, Q., Lin, S.C., Lamothe, B., Lu, M., Lo, Y.C., Hura, G., Zheng, L., Rich, R.L., Campos, A.D., Myszka, D.G., Lenardo, M.J., Darnay, B.G., Wu, H.(2009) Nat Struct Mol Biol 16: 658-666
- PubMed: 19465916
- DOI: https://doi.org/10.1038/nsmb.1605
- Primary Citation of Related Structures:
3HCS, 3HCT, 3HCU - PubMed Abstract:
Tumor necrosis factor (TNF) receptor-associated factor (TRAF)-6 mediates Lys63-linked polyubiquitination for NF-kappaB activation via its N-terminal RING and zinc finger domains. Here we report the crystal structures of TRAF6 and its complex with the ubiquitin-conjugating enzyme (E2) Ubc13. The RING and zinc fingers of TRAF6 assume a rigid, elongated structure. Interaction of TRAF6 with Ubc13 involves direct contacts of the RING and the preceding residues, and the first zinc finger has a structural role. Unexpectedly, this region of TRAF6 is dimeric both in the crystal and in solution, different from the trimeric C-terminal TRAF domain. Structure-based mutagenesis reveals that TRAF6 dimerization is crucial for polyubiquitin synthesis and autoubiquitination. Fluorescence resonance energy transfer analysis shows that TRAF6 dimerization induces higher-order oligomerization of full-length TRAF6. The mismatch of dimeric and trimeric symmetry may provide a mode of infinite oligomerization that facilitates ligand-dependent signal transduction of many immune receptors.
- Weill Medical College of Cornell University, New York, New York, USA.
Organizational Affiliation: