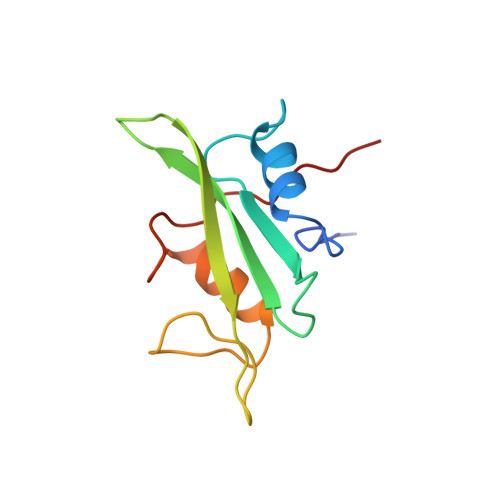
Sequential assignment and secondary structure determination for the Src homology 2 domain of hematopoietic cellular kinase.
Zhang, W., Smithgall, T.E., Gmeiner, W.H.(1997) FEBS Lett 406: 131-135
- PubMed: 9109402
- DOI: https://doi.org/10.1016/s0014-5793(97)00255-x
- Primary Citation of Related Structures:
3HCK - PubMed Abstract:
The hematopoietic cellular kinase (Hck) is a member of the Src family of non-receptor protein-tyrosine kinases and participates in signal transduction events regulating the growth, differentiation and function of phagocytes. The secondary structure of the SH2 domain for Hck was determined for a 13C/15N-enriched sample using multi-dimensional NMR spectroscopy. The secondary structure for the domain was determined from chemical shift indices [1H alpha, 13C alpha and 13C'], sequential NOEs [d(alphaN)(i, i+1) and d(NN)(i, i+1)], and 3J(alphaN) scalar coupling constants. The Hck SH2 domain consists of two alpha-helices and seven beta-strands. Complementary strands of beta-sheets were identified from long-range NOEs using a novel 3D, 13C/15N-edited HMQC-NOESY-(HCACO)NH experiment that correlated 1H alpha resonances between beta-strands. The secondary structure for Hck SH2 is similar to that predicted from the sequence alignment of the Src-family protein tyrosine kinases.
- The Eppley Institute for Research in Cancer and Allied Diseases, The University of Nebraska Medical Center, Omaha 68189-6805, USA.
Organizational Affiliation: