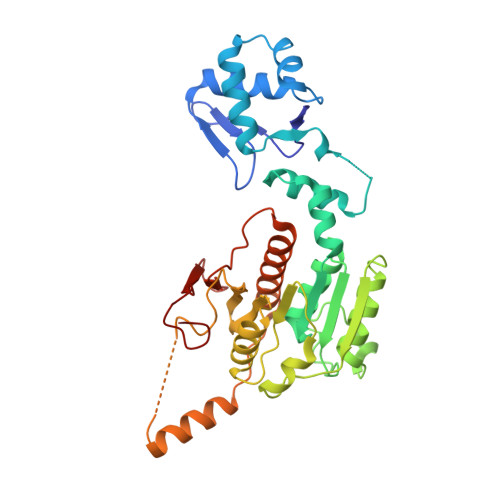
How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Regni, C.A., Roush, R.F., Miller, D.J., Nourse, A., Walsh, C.T., Schulman, B.A.(2009) EMBO J 28: 1953-1964
- PubMed: 19494832
- DOI: https://doi.org/10.1038/emboj.2009.146
- Primary Citation of Related Structures:
3H5A, 3H5N, 3H5R, 3H9G, 3H9J, 3H9Q - PubMed Abstract:
The 39-kDa Escherichia coli enzyme MccB catalyses a remarkable posttranslational modification of the MccA heptapeptide during the biosynthesis of microcin C7 (MccC7), a 'Trojan horse' antibiotic. The approximately 260-residue C-terminal region of MccB is homologous to ubiquitin-like protein (UBL) activating enzyme (E1) adenylation domains. Accordingly, MccB-catalysed C-terminal MccA-acyl-adenylation is reminiscent of the E1-catalysed activation reaction. However, unlike E1 substrates, which are UBLs with a C-terminal di-glycine sequence, MccB's substrate, MccA, is a short peptide with an essential C-terminal Asn. Furthermore, after an intramolecular rearrangement of MccA-acyl-adenylate, MccB catalyses a second, unique reaction, producing a stable phosphoramidate-linked analogue of acyl-adenylated aspartic acid. We report six-crystal structures of MccB in apo, substrate-, intermediate-, and inhibitor-bound forms. Structural and kinetic analyses reveal a novel-peptide clamping mechanism for MccB binding to heptapeptide substrates and a dynamic-active site for catalysing dual adenosine triphosphate-consuming reactions. The results provide insight into how a distinctive member of the E1 superfamily carries out two-step activation for generating the peptidyl-antibiotic MccC7.
- Department of Structural Biology, St Jude Children's Research Hospital, Memphis, TN 38105, USA.
Organizational Affiliation: