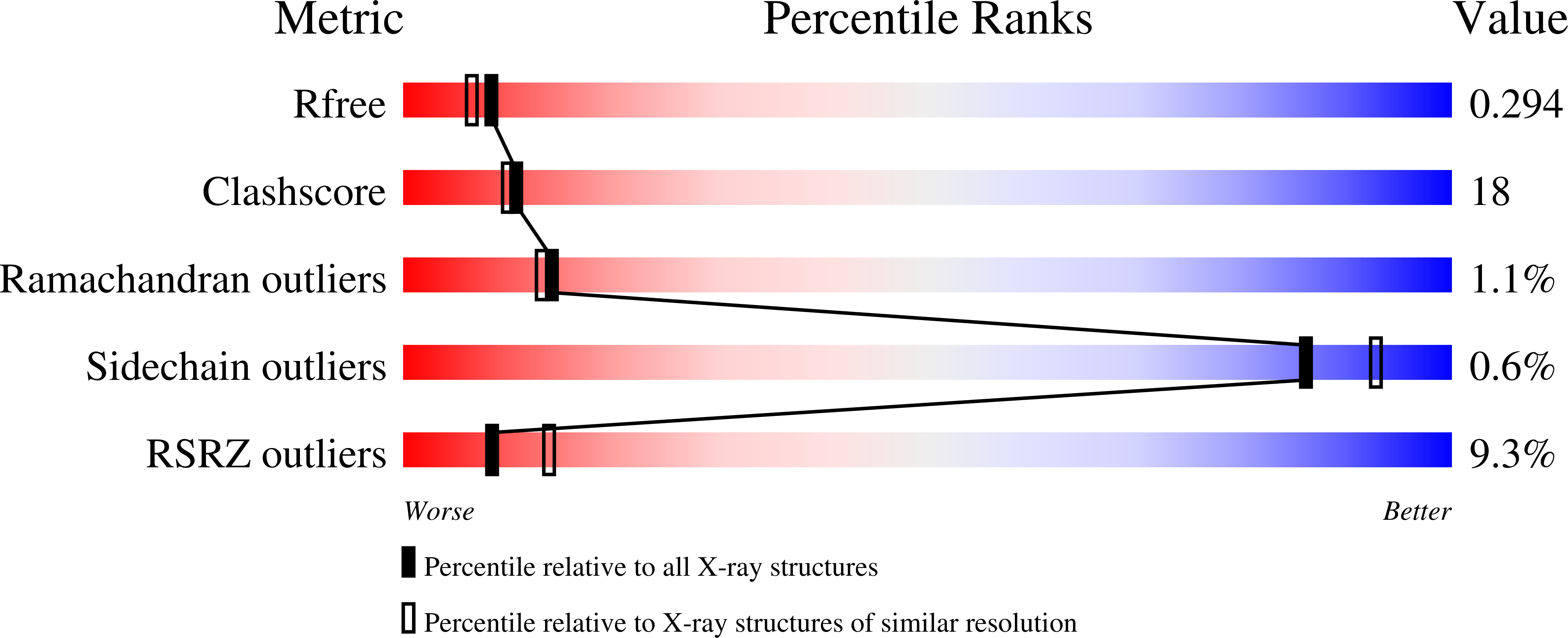
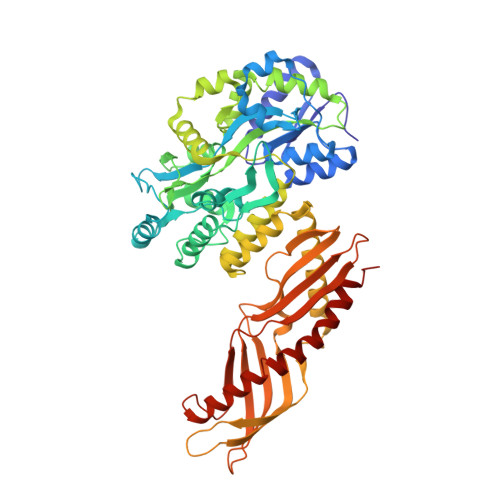
The structure of the dust mite allergen Der p 7 reveals similarities to innate immune proteins.
Mueller, G.A., Edwards, L.L., Aloor, J.J., Fessler, M.B., Glesner, J., Pomes, A., Chapman, M.D., London, R.E., Pedersen, L.C.(2010) J Allergy Clin Immunol 125: 909-917.e4
- PubMed: 20226507
- DOI: https://doi.org/10.1016/j.jaci.2009.12.016
- Primary Citation of Related Structures:
3H4Z - PubMed Abstract:
Sensitization to house dust mite allergens is strongly correlated with asthma. Der p 7 elicits strong IgE antibody and T-cell responses in patients with mite allergy. However, the structure and biological function of this important allergen are unknown. Allergen function might contribute to allergenicity, as shown for the protease activity of group 1 mite allergens and the interaction with the innate immune system by group 2 mite allergens. We sought to determine the crystal structure of Der p 7 and to investigate its biological function. X-ray crystallography was used to determine the Der p 7 structure. Nuclear magnetic resonance analysis and biochemical assays were used to examine the binding of Der p 7 to predicted ligands. Der p 7 has an elongated structure, with two 4-stranded antiparallel beta-sheets that wrap around a long C-terminal helix. The fold of Der p 7 is similar to that of LPS-binding protein (LBP), which interacts with Toll-like receptors after binding LPS and other bacterially derived lipid ligands. Nuclear magnetic resonance and biochemical assays indicate that Der p 7 does not bind LPS but binds with weak affinity to the bacterial lipopeptide polymyxin B in the predicted binding site of Der p 7. Der p 7 binds a bacterially derived lipid product, a common feature of some allergens. The finding that the group 7, as well as the group 2, mite allergens are structurally similar to different proteins in the Toll-like receptor pathway further strengthens the connections between dust mites, innate immunity, and allergy.
- Laboratory of Structural Biology, National Institute of Environmental Health Sciences, Research Triangle Park, NC, USA. mueller3@niehs.nih.gov
Organizational Affiliation: