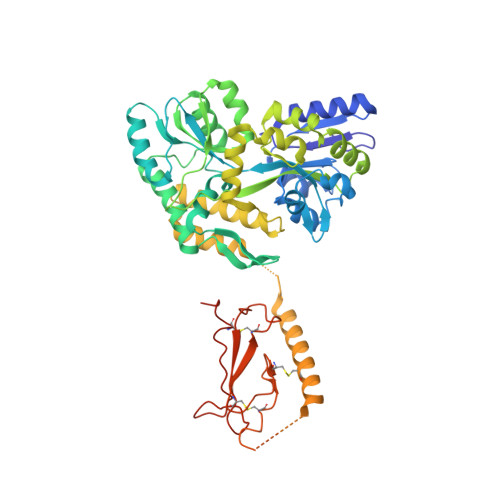
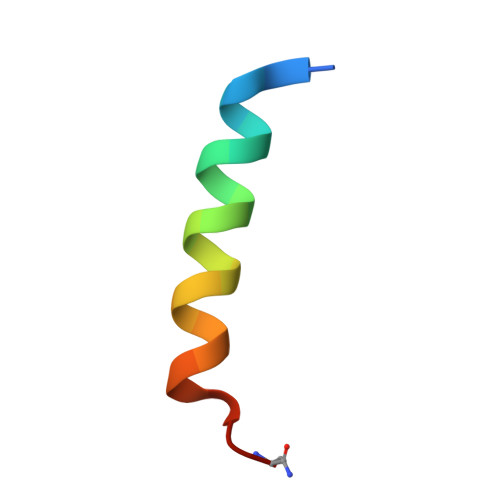
Structural basis for parathyroid hormone-related protein binding to the parathyroid hormone receptor and design of conformation-selective peptides.
Pioszak, A.A., Parker, N.R., Gardella, T.J., Xu, H.E.(2009) J Biological Chem 284: 28382-28391
- PubMed: 19674967
- DOI: https://doi.org/10.1074/jbc.M109.022905
- Primary Citation of Related Structures:
3H3G - PubMed Abstract:
Parathyroid hormone (PTH) and PTH-related protein (PTHrP) are two related peptides that control calcium/phosphate homeostasis and bone development, respectively, through activation of the PTH/PTHrP receptor (PTH1R), a class B G protein-coupled receptor. Both peptides hold clinical interest for their capacities to stimulate bone formation. PTH and PTHrP display different selectivity for two distinct PTH1R conformations, but how their binding to the receptor differs is unclear. The high resolution crystal structure of PTHrP bound to the extracellular domain (ECD) of PTH1R reveals that PTHrP binds as an amphipathic alpha-helix to the same hydrophobic groove in the ECD as occupied by PTH, but in contrast to a straight, continuous PTH helix, the PTHrP helix is gently curved and C-terminally "unwound." The receptor accommodates the altered binding modes by shifting the side chain conformations of two residues within the binding groove: Leu-41 and Ile-115, the former acting as a rotamer toggle switch to accommodate PTH/PTHrP sequence divergence, and the latter adapting to the PTHrP curvature. Binding studies performed with PTH/PTHrP hybrid ligands having reciprocal exchanges of residues involved in different contacts confirmed functional consequences for the altered interactions and enabled the design of altered PTH and PTHrP peptides that adopt the ECD-binding mode of the opposite peptide. Hybrid peptides that bound the ECD poorly were selective for the G protein-coupled PTH1R conformation. These results establish a molecular model for better understanding of how two biologically distinct ligands can act through a single receptor and provide a template for designing better PTH/PTHrP therapeutics.
- Laboratory of Structural Sciences, Van Andel Research Institute, Grand Rapids, Michigan 49503. Electronic address: augie.pioszak@vai.org.
Organizational Affiliation: