The design and optimization of a series of 2-(pyridin-2-yl)-1H-benzimidazole compounds as allosteric glucokinase activators.
Takahashi, K., Hashimoto, N., Nakama, C., Kamata, K., Sasaki, K., Yoshimoto, R., Ohyama, S., Hosaka, H., Maruki, H., Nagata, Y., Eiki, J., Nishimura, T.(2009) Bioorg Med Chem 17: 7042-7051
- PubMed: 19736020
- DOI: https://doi.org/10.1016/j.bmc.2009.05.037
- Primary Citation of Related Structures:
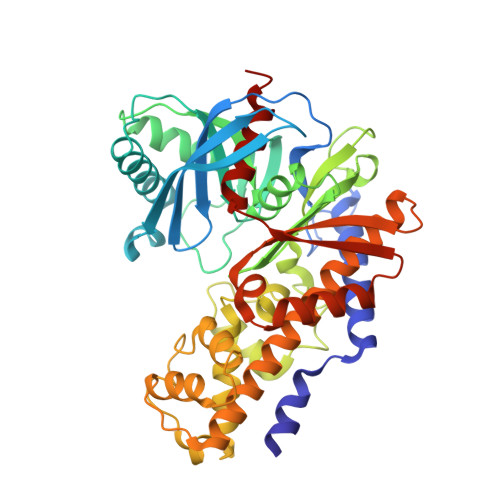
3H1V - PubMed Abstract:
The optimization of a series of benzimidazole glucokinase activators is described. We identified a novel and potent achiral benzimidazole derivative as an allosteric GK activator. This activator was designed and synthesized via removal of the chiral center of the lead compound, 6-(N-acylpyrrolidin-2-yl)benzimidazole. The activator exhibited good PK profiles in rats and dogs, and significant hypoglycemic efficacy at 1 mg/kg po dosing in a rat OGTT model. The binding site and binding mode of the benzimidazole class of GKA with GK protein was confirmed by X-ray crystallographic analysis.
- Banyu Tsukuba Research Institute, Banyu Pharmaceutical Co., Ltd, 3 Okubo, Tsukuba, Ibaraki 300-2611, Japan. g94e079@ruri.waseda.jp
Organizational Affiliation: