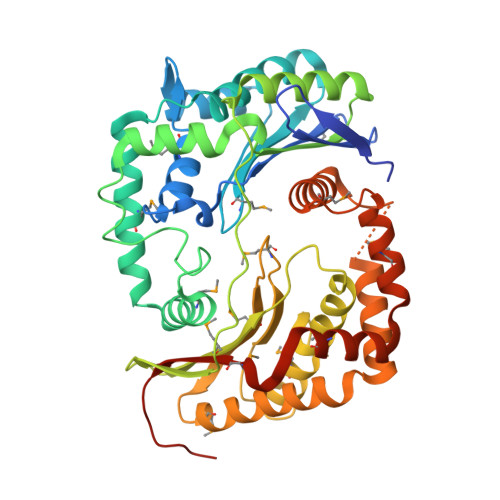
Crystal structure of peptidase M16 inactive domain from Pseudomonas fluorescens. Northeast Structural Genomics target PlR293L
Seetharaman, J., Chen, Y., Fang, F., Xiao, R., Everett, J.K., Acton, T.B., Rost, B., Montelione, G.T., Tong, L., Hunt, J.F.To be published.