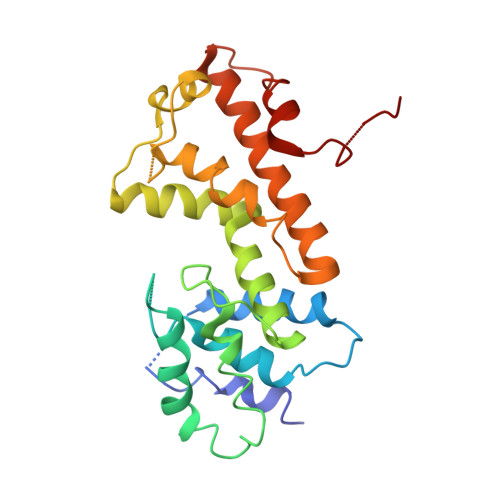
Crystal structure of a metal-dependent phosphohydrolase with conserved HD domain (yedJ) from Escherichia coli in complex with nickel ions. Northeast Structural Genomics Consortium Target ER63
Forouhar, F., Abashidze, M., Seetharaman, J., Janjua, J., Xiao, R., Cunningham, K., Ma, L., Zhao, L., Everett, J.K., Nair, R., Acton, T.B., Rost, B., Montelione, G.T., Hunt, J.F., Tong, L.To be published.