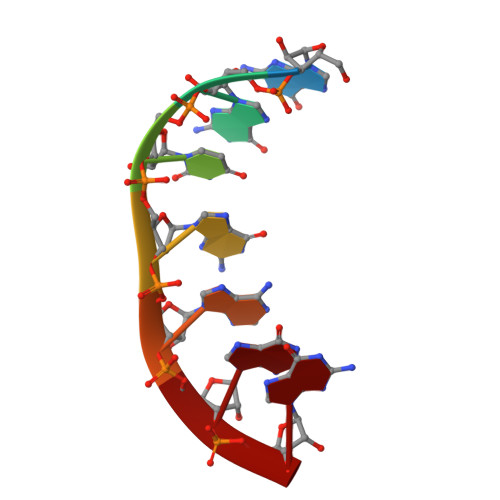
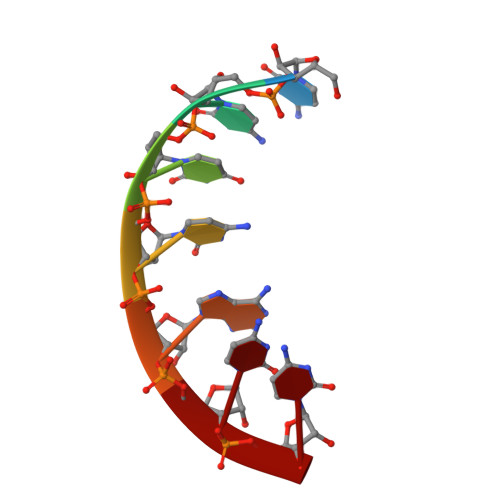
The 1.2A crystal structure of an E. coli tRNASer)acceptor stem microhelix reveals two magnesium binding sites.
Eichert, A., Furste, J.P., Schreiber, A., Perbandt, M., Betzel, C., Erdmann, V.A., Forster, C.(2009) Biochem Biophys Res Commun 386: 368-373
- PubMed: 19527687
- DOI: https://doi.org/10.1016/j.bbrc.2009.06.048
- Primary Citation of Related Structures:
3GVN - PubMed Abstract:
tRNA identity elements assure the correct aminoacylation of tRNAs by the cognate aminoacyl-tRNA synthetases. tRNA(Ser) belongs to the so-called class II system, in which the identity elements are rather simple and are mostly located in the acceptor stem region, in contrast to 'class I', where tRNA determinants are more complex and are located within different regions of the tRNA. The structure of an Escherichia coli tRNA(Ser) acceptor stem microhelix was solved by high resolution X-ray structure analysis. The RNA crystallizes in the space group C2, with one molecule per asymmetric unit and with the cell constants a=35.79, b=39.13, c=31.37A, and beta=111.1 degrees . A defined hydration pattern of 97 water molecules surrounds the tRNA(Ser) acceptor stem microhelix. Additionally, two magnesium binding sites were detected in the tRNA(Ser) aminoacyl stem.
- Institute of Chemistry and Biochemistry, Free University Berlin, Thielallee 63, 14195 Berlin, Germany.
Organizational Affiliation: