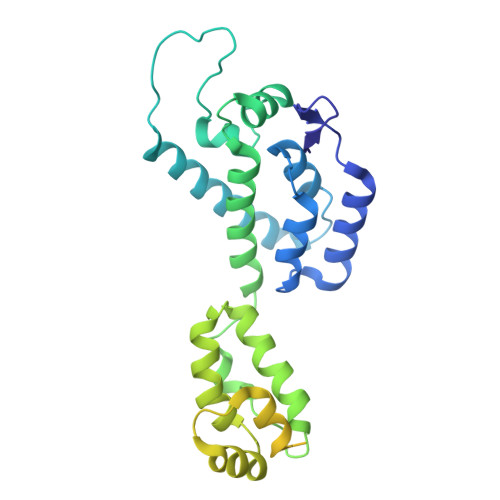
X-ray structures of the hexameric building block of the HIV capsid.
Pornillos, O., Ganser-Pornillos, B.K., Kelly, B.N., Hua, Y., Whitby, F.G., Stout, C.D., Sundquist, W.I., Hill, C.P., Yeager, M.(2009) Cell 137: 1282-1292
- PubMed: 19523676
- DOI: https://doi.org/10.1016/j.cell.2009.04.063
- Primary Citation of Related Structures:
3GV2, 3H47, 3H4E - PubMed Abstract:
The mature capsids of HIV and other retroviruses organize and package the viral genome and its associated enzymes for delivery into host cells. The HIV capsid is a fullerene cone: a variably curved, closed shell composed of approximately 250 hexamers and exactly 12 pentamers of the viral CA protein. We devised methods for isolating soluble, assembly-competent CA hexamers and derived four crystallographically independent models that define the structure of this capsid assembly unit at atomic resolution. A ring of six CA N-terminal domains form an apparently rigid core, surrounded by an outer ring of C-terminal domains. Mobility of the outer ring appears to be an underlying mechanism for generating the variably curved lattice in authentic capsids. Hexamer-stabilizing interfaces are highly hydrated, and this property may be key to the formation of quasi-equivalent interactions within hexamers and pentamers. The structures also clarify the molecular basis for capsid assembly inhibition and should facilitate structure-based drug design strategies.
- Department of Cell Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: