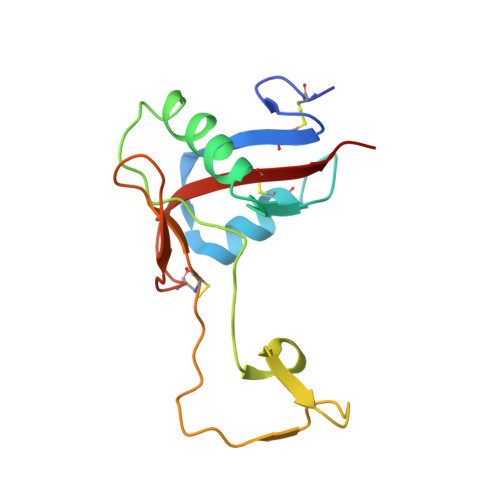
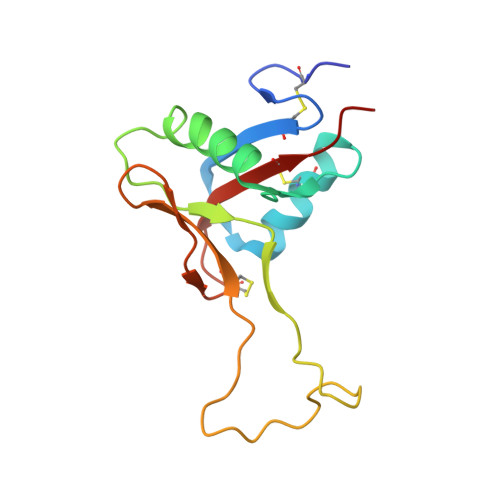
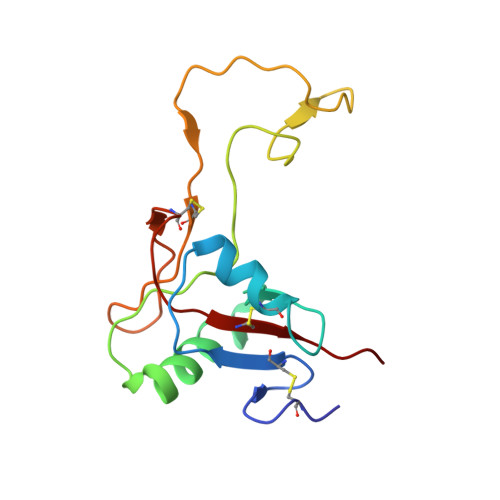
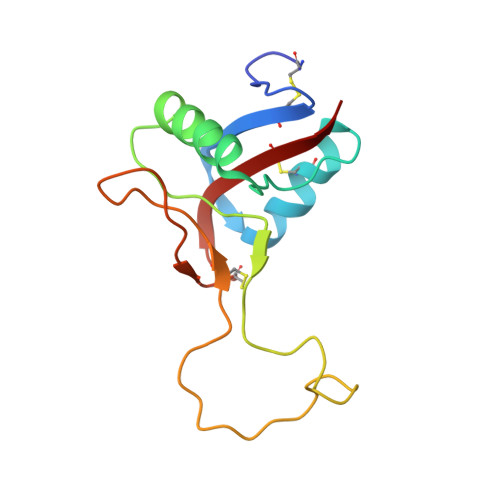
The alpha2beta1 integrin-specific antagonist rhodocetin is a cruciform, heterotetrameric molecule
Eble, J.A., Niland, S., Bracht, T., Mormann, M., Peter-Katalinic, J., Pohlentz, G., Stetefeld, J.(2009) FASEB J 23: 2917-2927
- PubMed: 19369383
- DOI: https://doi.org/10.1096/fj.08-126763
- Primary Citation of Related Structures:
3GPR - PubMed Abstract:
The integrin alpha2beta1 plays an important role in various pathophysiological processes, such as thrombosis, wound healing, inflammation, and metastasis. Rhodocetin, a constituent of the venom of the hemorrhagic Malayan pit viper (Calloselasma rhodostoma), is a specific alpha2beta1 integrin antagonist. To understand its molecular mode of action, its structure was studied by crystallography. Its quaternary structure in solution was also analyzed biochemically. Two novel subunits of rhodocetin were sequenced by mass spectrometry. Their integrin binding was measured by protein interaction ELISAs. Rhodocetin is a C-type lectin-like protein (CLP) consisting of four homologous, yet distinct, subunits, alpha, beta, gamma, and delta, the latter two of which have been unknown to date. With their CLP folds and loop-swapping motifs, the subunits alpha, beta and gamma, delta form two heterodimeric pairs. Uniquely, they arrange orthogonally and shape a cruciform molecule. Bearing a single unpaired cysteine residue, rhodocetin can only form covalent supramolecular complexes with a maximum aggregation number of 2, unlike many heterodimeric CLPs. Being the first heterotetrameric CLP to be crystallized, rhodocetin provides not only the prototypic molecular structure for heterotetrameric CLPs, but also a lead structure for pharmaceutical alpha2beta1 integrin antagonists.
- Center for Molecular Medicine, Department of Vascular Matrix Biology, Excellence Cluster Cardio-Pulmonary System, Frankfurt University Hospital, Frankfurt, Germany. eble@med.uni-frankfurt.de
Organizational Affiliation: