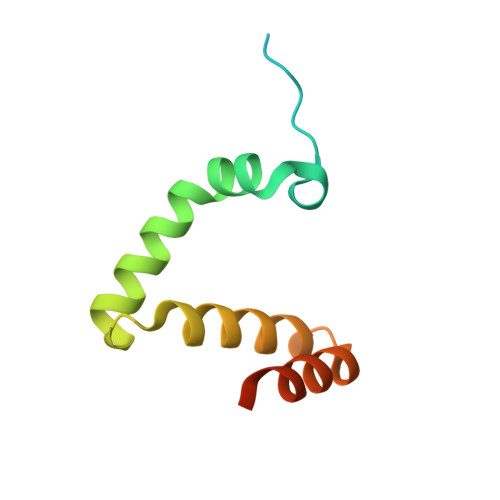
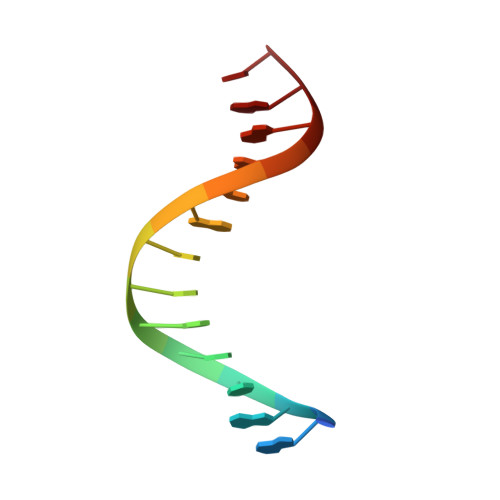
Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis.
Yin, F.F., Bailey, S., Innis, C.A., Ciubotaru, M., Kamtekar, S., Steitz, T.A., Schatz, D.G.(2009) Nat Struct Mol Biol 16: 499-508
- PubMed: 19396172
- DOI: https://doi.org/10.1038/nsmb.1593
- Primary Citation of Related Structures:
3GNA, 3GNB - PubMed Abstract:
The products of recombination-activating genes RAG1 and RAG2 mediate the assembly of antigen receptor genes during lymphocyte development in a process known as V(D)J recombination. Lack of structural information for the RAG proteins has hindered mechanistic studies of this reaction. We report here the crystal structure of an essential DNA binding domain of the RAG1 catalytic core bound to its nonamer DNA recognition motif. The RAG1 nonamer binding domain (NBD) forms a tightly interwoven dimer that binds and synapses two nonamer elements, with each NBD making contact with both DNA molecules. Biochemical and biophysical experiments confirm that the two nonamers are in close proximity in the RAG1/2-DNA synaptic complex and demonstrate the functional importance of the protein-DNA contacts revealed in the structure. These findings reveal a previously unsuspected function for the NBD in DNA synapsis and have implications for the regulation of DNA binding and cleavage by RAG1 and RAG2.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut, USA.
Organizational Affiliation: