Crystal structure of ATPase domain of Ssb1 chaperone, member of the HSP70 family from Saccharomyces cerevisiae.
Osipiuk, J., Li, H., Bargassa, M., Sahi, C., Craig, E.A., Joachimiak, A.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Heat shock protein SSB1 | 387 | Saccharomyces cerevisiae | Mutation(s): 0 Gene Names: SSB1, YDL229W, YG101 EC: 3.6.4.10 | 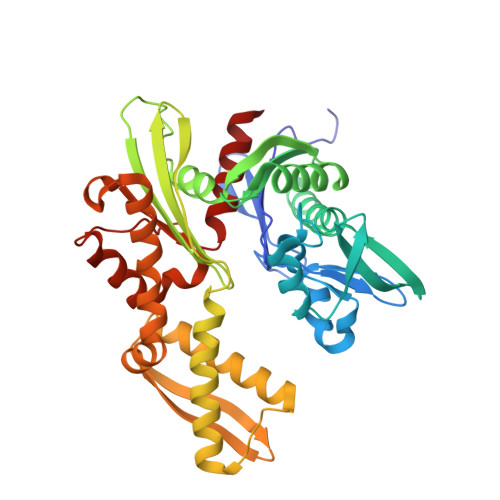 | |
UniProt | |||||
Find proteins for P11484 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P11484 Go to UniProtKB: P11484 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P11484 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GOL Query on GOL | M [auth B] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| CL Query on CL | E [auth A] F [auth A] G [auth A] H [auth A] J [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| MG Query on MG | C [auth A], D [auth A], I [auth B] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 76.543 | α = 90 |
| b = 54.504 | β = 104.36 |
| c = 90.623 | γ = 90 |
| Software Name | Purpose |
|---|---|
| DENZO | data reduction |
| SCALEPACK | data scaling |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| SBC-Collect | data collection |
| HKL-2000 | data reduction |
| MOLREP | phasing |