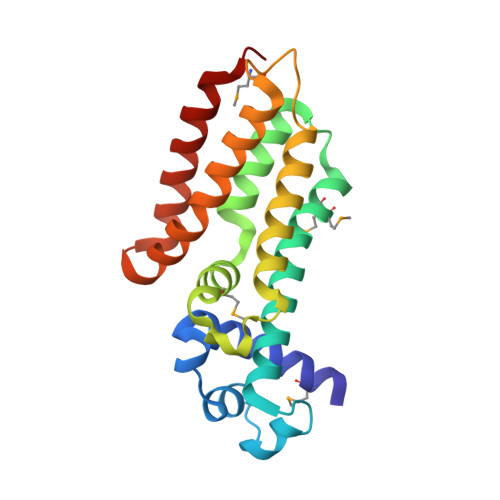
Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family
Anderson, S.M., Brunzelle, J.S., Wawrzak, Z., Skarina, T., Papazisi, L., Anderson, W.F., Savchenko, A., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.