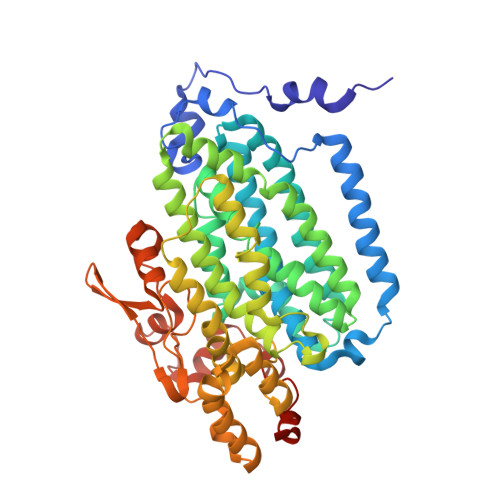
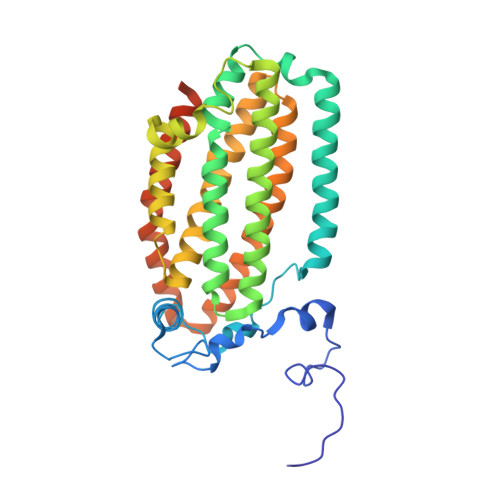
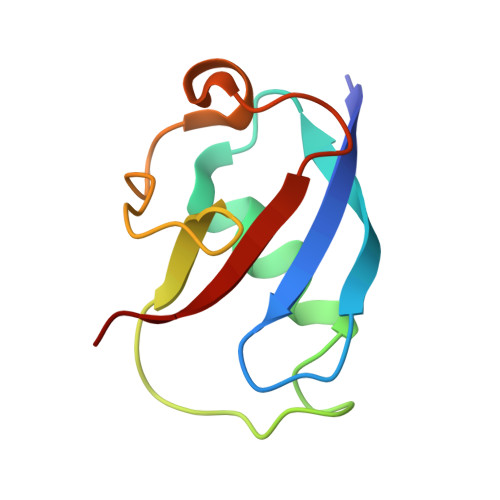
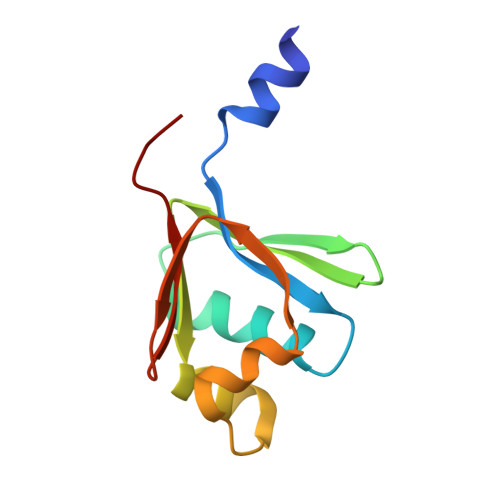
Role for threonine 201 in the catalytic cycle of the soluble diiron hydroxylase toluene 4-monooxygenase.
Elsen, N.L., Bailey, L.J., Hauser, A.D., Fox, B.G.(2009) Biochemistry 48: 3838-3846
- PubMed: 19290655
- DOI: https://doi.org/10.1021/bi900144a
- Primary Citation of Related Structures:
3GE3, 3GE8 - PubMed Abstract:
The active site residue Thr-201 in toluene 4-monooxygenase hydroxylase (T4moH) has a structural counterpart in the active sites of all diiron monooxygenases. Thus, our previous finding that mutation of this residue to Ala, Gly, or Ser had no impact on steady-state catalysis or coupling was surprising. In this work, we provide kinetic, biochemical, and structural evidence that one role of Thr-201 may be to stabilize a peroxo-level intermediate during enzyme catalysis. During reactions in the absence of substrate, T201 T4moH slowly consumed O(2) but only a negligible amount of H(2)O(2) was released. In contrast, T201A T4moH gave stoichometric release of H(2)O(2) during reaction in the absence of substrate. Both enzyme isoforms were tightly coupled during steady-state catalysis with saturating toluene and other optimal substrates and exhibited near-identical kinetic parameters. However, rapid mix single-turnover studies showed that T201A T4moH had a faster first-order rate constant for product formation than T201 T4moH did. Comparison of X-ray crystal structures of resting and reduced T201A T4moH in complex with T4moD with comparable structures of T201 T4moHD revealed changes in the positions of several key active site residues relative to the comparable structures of T201 T4moH with T4moD. This combination of catalytic and structural studies offers important new insight into the role of the role of conserved Thr-201, and its contributions to the catalytic reaction cycle.
- Department of Biochemistry, College of Agricultural and Life Sciences, University of Wisconsin, Madison, Wisconsin 53706-1544, USA.
Organizational Affiliation: