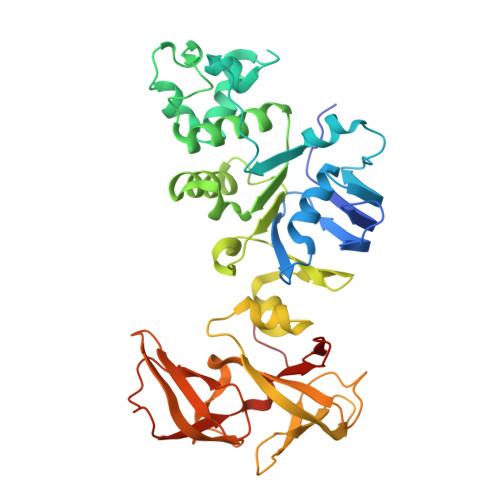
Crystal structure of human NBD2 complexed with N6-Phenylethyl-ATP (P-ATP)
Atwell, S., Antonysamy, S., Guggino, W.B., Conners, K., Emtage, S., Gheyi, T., Hunt, J.F., Lewis, H.A., Lu, F., Sauder, J.M., Weber, P.C., Wetmore, D., Zhao, X.To be published.