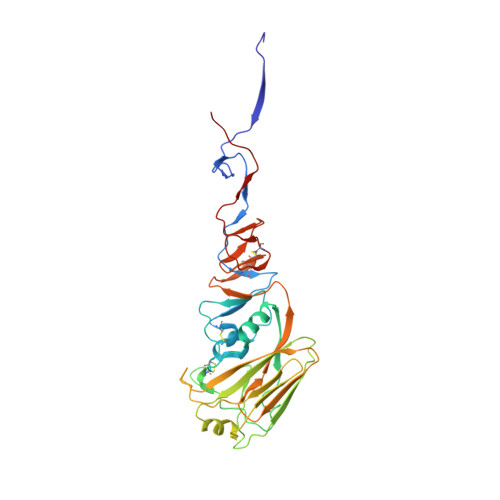
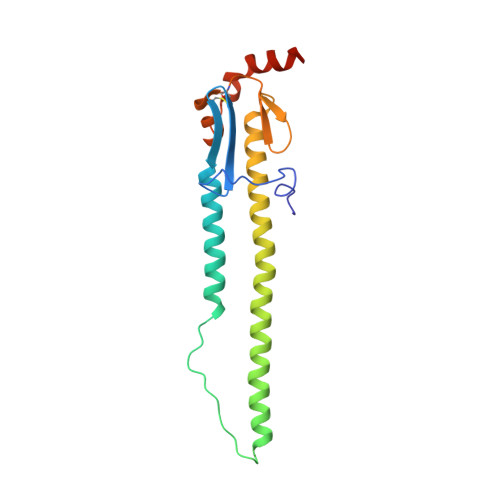
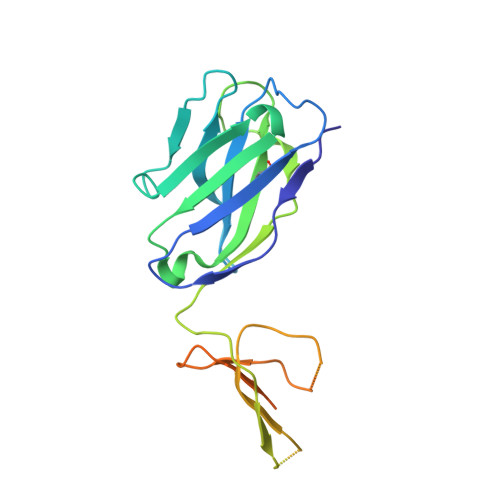
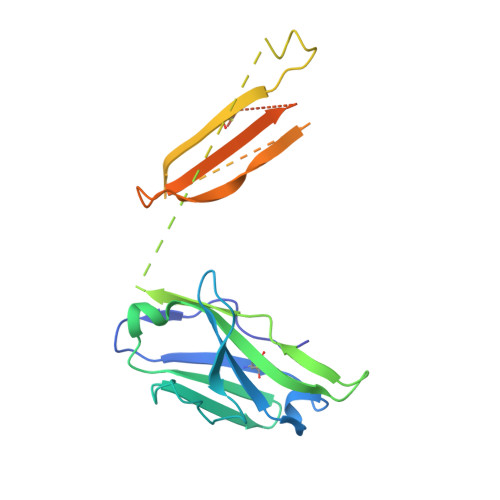
Antibody recognition of a highly conserved influenza virus epitope.
Ekiert, D.C., Bhabha, G., Elsliger, M.A., Friesen, R.H., Jongeneelen, M., Throsby, M., Goudsmit, J., Wilson, I.A.(2009) Science 324: 246-251
- PubMed: 19251591
- DOI: https://doi.org/10.1126/science.1171491
- Primary Citation of Related Structures:
3GBM, 3GBN - PubMed Abstract:
Influenza virus presents an important and persistent threat to public health worldwide, and current vaccines provide immunity to viral isolates similar to the vaccine strain. High-affinity antibodies against a conserved epitope could provide immunity to the diverse influenza subtypes and protection against future pandemic viruses. Cocrystal structures were determined at 2.2 and 2.7 angstrom resolutions for broadly neutralizing human antibody CR6261 Fab in complexes with the major surface antigen (hemagglutinin, HA) from viruses responsible for the 1918 H1N1 influenza pandemic and a recent lethal case of H5N1 avian influenza. In contrast to other structurally characterized influenza antibodies, CR6261 recognizes a highly conserved helical region in the membrane-proximal stem of HA1 and HA2. The antibody neutralizes the virus by blocking conformational rearrangements associated with membrane fusion. The CR6261 epitope identified here should accelerate the design and implementation of improved vaccines that can elicit CR6261-like antibodies, as well as antibody-based therapies for the treatment of influenza.
- Department of Molecular Biology, Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA.
Organizational Affiliation: