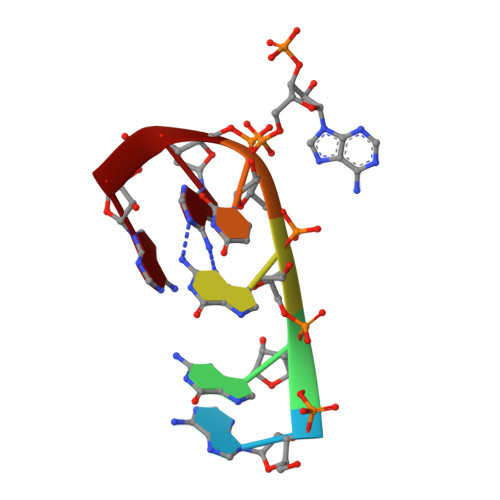
The zinc fingers of the SR-like protein ZRANB2 are single-stranded RNA-binding domains that recognize 5' splice site-like sequences
Loughlin, F.E., Mansfield, R.E., Vaz, P.M., McGrath, A.P., Setiyaputra, S., Gamsjaeger, R., Chen, E.S., Morris, B.J., Guss, J.M., Mackay, J.P.(2009) Proc Natl Acad Sci U S A 106: 5581-5586
- PubMed: 19304800
- DOI: https://doi.org/10.1073/pnas.0802466106
- Primary Citation of Related Structures:
3G9Y - PubMed Abstract:
The alternative splicing of mRNA is a critical process in higher eukaryotes that generates substantial proteomic diversity. Many of the proteins that are essential to this process contain arginine/serine-rich (RS) domains. ZRANB2 is a widely-expressed and highly-conserved RS-domain protein that can regulate alternative splicing but lacks canonical RNA-binding domains. Instead, it contains 2 RanBP2-type zinc finger (ZnF) domains. We demonstrate that these ZnFs recognize ssRNA with high affinity and specificity. Each ZnF binds to a single AGGUAA motif and the 2 domains combine to recognize AGGUAA(N(x))AGGUAA double sites, suggesting that ZRANB2 regulates alternative splicing via a direct interaction with pre-mRNA at sites that resemble the consensus 5' splice site. We show using X-ray crystallography that recognition of an AGGUAA motif by a single ZnF is dominated by side-chain hydrogen bonds to the bases and formation of a guanine-tryptophan-guanine "ladder." A number of other human proteins that function in RNA processing also contain RanBP2 ZnFs in which the RNA-binding residues of ZRANB2 are conserved. The ZnFs of ZRANB2 therefore define another class of RNA-binding domain, advancing our understanding of RNA recognition and emphasizing the versatility of ZnF domains in molecular recognition.
- School of Molecular and Microbial Biosciences, University of Sydney, Sydney NSW 2006, Australia.
Organizational Affiliation: