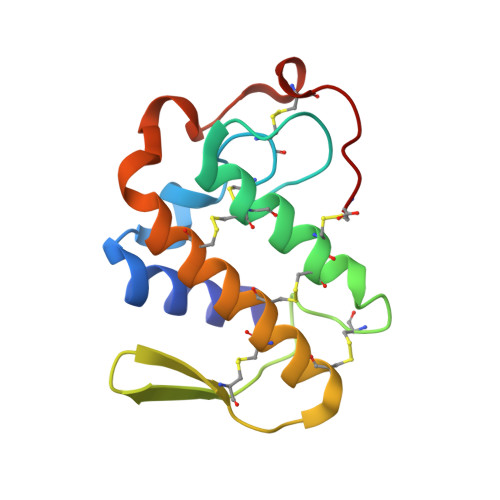
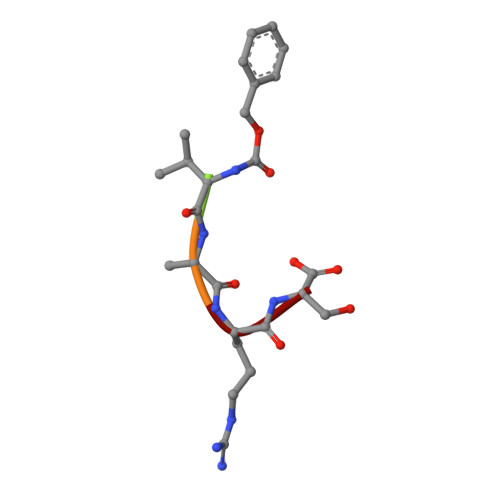
Crystal Structure of the Complex Formed between a Group II Phospholipase A2 and Designed Peptide Inhibitor Carbobenzoxy-Dehydro-Val-Ala-Arg-Ser at 1.2 A Resolution
Singh, N., Kaur, P., Prem Kumar, R., Somvanshi, R.K., Perband, M., Betzel, C., Dey, S., Sharma, S., Singh, T.P.To be published.