DNA Synthesis across an Abasic Lesion by Human DNA Polymerase iota
Nair, D.T., Johnson, R.E., Prakash, L., Prakash, S., Aggarwal, A.K.(2009) Structure 17: 530-537
- PubMed: 19368886
- DOI: https://doi.org/10.1016/j.str.2009.02.015
- Primary Citation of Related Structures:
3G6V, 3G6X, 3G6Y - PubMed Abstract:
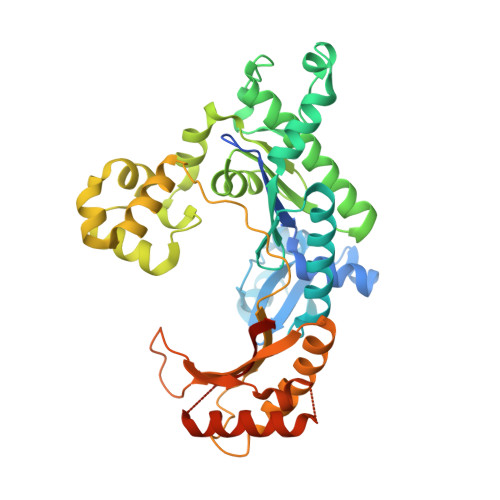
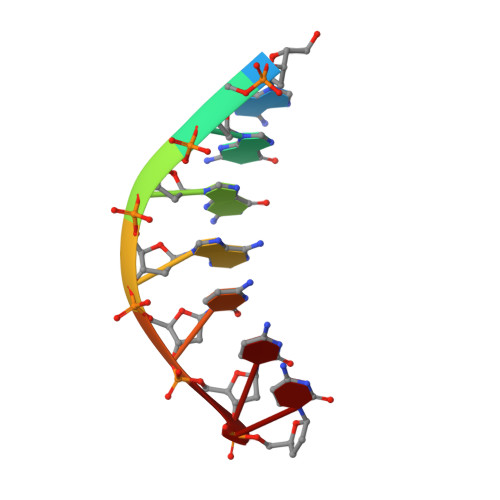
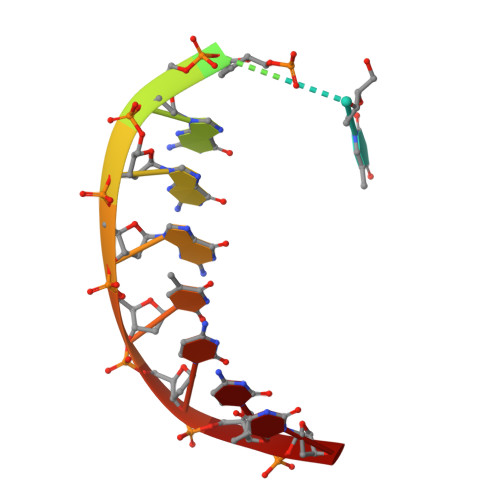
Abasic sites are among the most abundant DNA lesions formed in human cells, and they present a strong block to replication. DNA polymerase iota (Poliota) is one of the few DNA Pols that does not follow the A-rule opposite an abasic site. We present here three structures of human Poliota in complex with DNAs containing an abasic lesion and dGTP, dTTP, or dATP as the incoming nucleotide. The structures reveal a mechanism of translesion synthesis across an abasic lesion that differs from that in other Pols. Both the abasic lesion and the incoming dNTPs are intrahelical and are closely apposed across a constricted active site cleft. The dNTPs partake in distinct networks of hydrogen bonds in the "void" opposite the lesion. These different patterns of hydrogen bonds, as well as stacking interactions, may underlie Poliota's small preference for insertion of dGTP over other nucleotides opposite this common lesion.
- Department of Structural and Chemical Biology, Mount Sinai School of Medicine, New York, NY 10029, USA.
Organizational Affiliation: