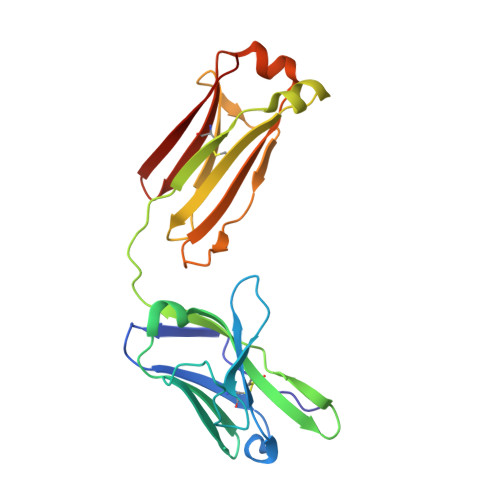
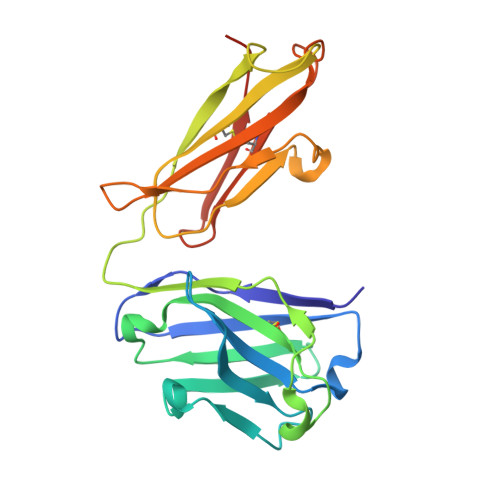
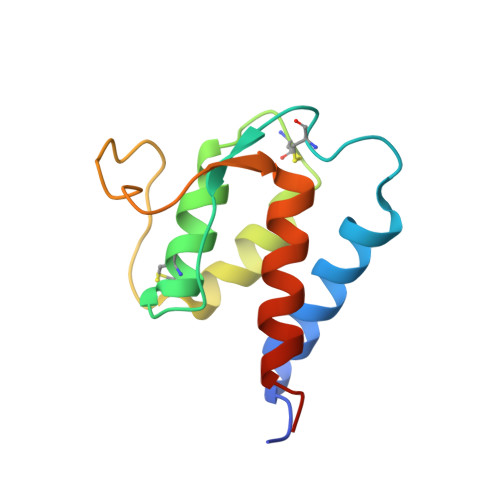
Epitope mapping of anti-interleukin-13 neutralizing antibody CNTO607.
Teplyakov, A., Obmolova, G., Wu, S.J., Luo, J., Kang, J., O'Neil, K., Gilliland, G.L.(2009) J Mol Biology 389: 115-123
- PubMed: 19361524
- DOI: https://doi.org/10.1016/j.jmb.2009.03.076
- Primary Citation of Related Structures:
3G6A, 3G6D - PubMed Abstract:
CNTO607 is a neutralizing anti-interleukin-13 (IL-13) human monoclonal antibody obtained from a phage display library. To determine how this antibody inhibits the biological effect of IL-13, we determined the binding epitope by X-ray crystallography. The crystal structure of the complex between CNTO607 Fab and IL-13 reveals the antibody epitope at the surface formed by helices A and D of IL-13. This epitope overlaps with the IL-4Ralpha/IL-13Ralpha1 receptor-binding site, which explains the neutralizing effect of CNTO607. The extensive antibody interface covers an area of 1000 A(2), which is consistent with the high binding affinity. The key features of the interface are the charge and shape complementarity of the molecules that include two hydrophobic pockets on IL-13 that accommodate Phe32 [complementarity-determining region (CDR) L2] and Trp100a (CDR H3) and a number of salt bridges between basic residues of IL-13 and acidic residues of the antibody. Comparison with the structure of the free Fab shows that the CDR residues do not change their conformation upon complex formation, with the exception of two residues in CDR H3, Trp100a and Asp100b, which change rotamer conformations. To evaluate the relative contribution of the epitope residues to CNTO607 binding, we performed alanine-scanning mutagenesis of the A-D region of IL-13. This study confirmed the primary role of electrostatic interactions for antigen recognition.
- Centocor R&D, Inc., Radnor, PA 19087, USA. ateplyak@its.jnj.com
Organizational Affiliation: