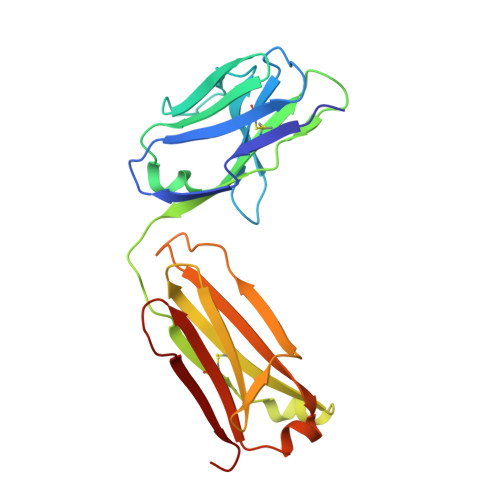
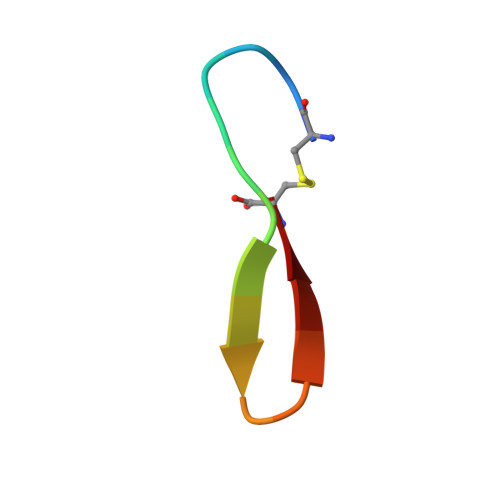
Antibodies specifically targeting a locally misfolded region of tumor associated EGFR
Garrett, T.P.J., Burgess, A.W., Gan, H.K., Luwor, R.B., Cartwright, G., Walker, F., Orchard, S.G., Clayton, A.H.A., Nice, E.C., Rothacker, J., Catimel, B., Cavenee, W.K., Old, L.J., Stockert, E., Ritter, G., Adams, T.E., Hoyne, P.A., Wittrup, D., Chao, G., Cochran, J.R., Luo, C., Lou, M., Huyton, T., Xu, Y., Fairlie, W.D., Yao, S., Scott, A.M., Johns, T.G.(2009) Proc Natl Acad Sci U S A 106: 5082-5087
- PubMed: 19289842
- DOI: https://doi.org/10.1073/pnas.0811559106
- Primary Citation of Related Structures:
3G5V, 3G5X, 3G5Y, 3G5Z - PubMed Abstract:
Epidermal Growth Factor Receptor (EGFR) is involved in stimulating the growth of many human tumors, but the success of therapeutic agents has been limited in part by interference from the EGFR on normal tissues. Previously, we reported an antibody (mab806) against a truncated form of EGFR found commonly in gliomas. Remarkably, it also recognizes full-length EGFR on tumor cells but not on normal cells. However, the mechanism for this activity was unclear. Crystallographic structures for Fab:EGFR(287-302) complexes of mAb806 (and a second, related antibody, mAb175) show that this peptide epitope adopts conformations similar to those found in the wtEGFR. However, in both conformations observed for wtEGFR, tethered and untethered, antibody binding would be prohibited by significant steric clashes with the CR1 domain. Thus, these antibodies must recognize a cryptic epitope in EGFR. Structurally, it appeared that breaking the disulfide bond preceding the epitope might allow the CR1 domain to open up sufficiently for antibody binding. The EGFR(C271A/C283A) mutant not only binds mAb806, but binds with 1:1 stoichiometry, which is significantly greater than wtEGFR binding. Although mAb806 and mAb175 decrease tumor growth in xenografts displaying mutant, overexpressed, or autocrine stimulated EGFR, neither antibody inhibits the in vitro growth of cells expressing wtEGFR. In contrast, mAb806 completely inhibits the ligand-associated stimulation of cells expressing EGFR(C271A/C283A). Clearly, the binding of mAb806 and mAb175 to the wtEGFR requires the epitope to be exposed either during receptor activation, mutation, or overexpression. This mechanism suggests the possibility of generating antibodies to target other wild-type receptors on tumor cells.
- Walter and Eliza Hall Institute of Medical Research, Parkville 3050, Australia.
Organizational Affiliation: