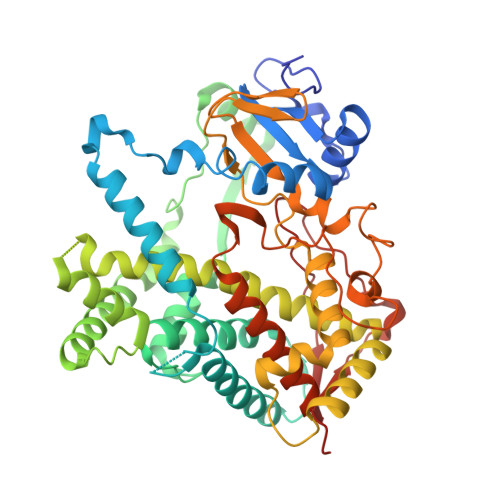
Crystal structures of cytochrome P450 2B4 in complex with the inhibitor 1-biphenyl-4-methyl-1H-imidazole: ligand-induced structural response through alpha-helical repositioning.
Gay, S.C., Sun, L., Maekawa, K., Halpert, J.R., Stout, C.D.(2009) Biochemistry 48: 4762-4771
- PubMed: 19397311
- DOI: https://doi.org/10.1021/bi9003765
- Primary Citation of Related Structures:
3G5N, 3G93 - PubMed Abstract:
Two different ligand occupancy structures of cytochrome P450 2B4 (CYP2B4) in complex with 1-biphenyl-4-methyl-1H-imidazole (1-PBI) have been determined by X-ray crystallography. 1-PBI belongs to a series of tight binding, imidazole-based CYP2B4 inhibitors. 1-PBI binding to CYP2B4 yields a type II spectrum with a K(s) value of 0.23 microM and inhibits enzyme activity with an IC(50) value of 0.035 microM. Previous CYP2B4 structures have shown a large degree of structural movement in response to ligand size. With two phenyl rings, 1-PBI is larger than 1-(4-chlorophenyl)imidazole (1-CPI) and 4-(4-chlorophenyl)imidazole (4-CPI) but smaller than bifonazole, which is branched and contains three phenyl rings. The CYP2B4-1-PBI complex is a structural intermediate to the closed CPI and the open bifonazole structures. The B/C-loop reorganizes itself to include two short partial helices while closing one side of the active site. The F-G-helix cassette pivots over the I-helix in direct response to the size of the ligand in the active site. A cluster of Phe residues at the fulcrum of this pivot point allows for dramatic repositioning of the cassette with only a relatively small amount of secondary structure rearrangement. Comparisons of ligand-bound CYP2B4 structures reveal trends in plastic region mobility that could allow for predictions of their position in future structures based on ligand shape and size.
- Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California at San Diego, La Jolla, California 92093, USA. scgay@ucsd.edu
Organizational Affiliation: