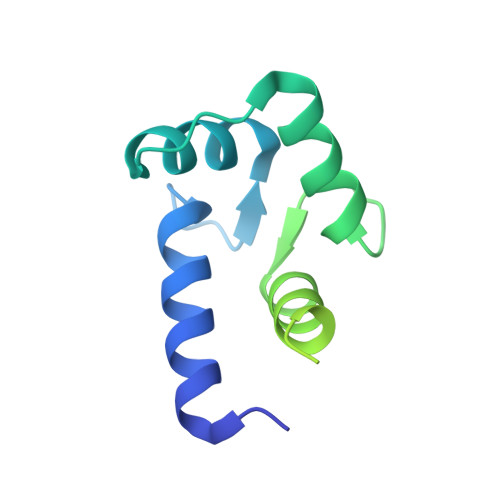
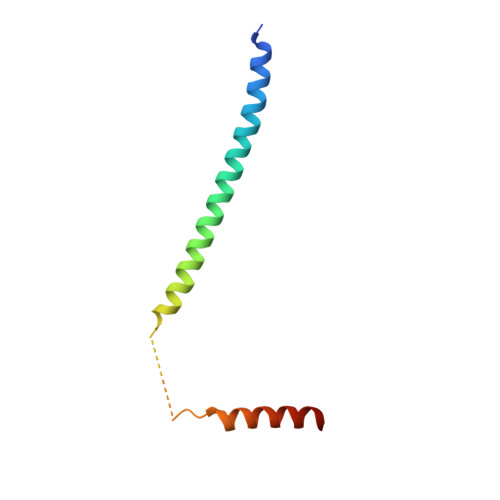
Crystal structure of dimeric cardiac L-type calcium channel regulatory domains bridged by Ca2+{middle dot}calmodulins.
Fallon, J.L., Baker, M.R., Xiong, L., Loy, R.E., Yang, G., Dirksen, R.T., Hamilton, S.L., Quiocho, F.A.(2009) Proc Natl Acad Sci U S A 106: 5135-5140
- PubMed: 19279214
- DOI: https://doi.org/10.1073/pnas.0807487106
- Primary Citation of Related Structures:
3G43 - PubMed Abstract:
Voltage-dependent calcium channels (Ca(V)) open in response to changes in membrane potential, but their activity is modulated by Ca(2+) binding to calmodulin (CaM). Structural studies of this family of channels have focused on CaM bound to the IQ motif; however, the minimal differences between structures cannot adequately describe CaM's role in the regulation of these channels. We report a unique crystal structure of a 77-residue fragment of the Ca(V)1.2 alpha(1) subunit carboxyl terminus, which includes a tandem of the pre-IQ and IQ domains, in complex with Ca(2+).CaM in 2 distinct binding modes. The structure of the Ca(V)1.2 fragment is an unusual dimer of 2 coiled-coiled pre-IQ regions bridged by 2 Ca(2+).CaMs interacting with the pre-IQ regions and a canonical Ca(V)1-IQ-Ca(2+).CaM complex. Native Ca(V)1.2 channels are shown to be a mixture of monomers/dimers and a point mutation in the pre-IQ region predicted to abolish the coiled-coil structure significantly reduces Ca(2+)-dependent inactivation of heterologously expressed Ca(V)1.2 channels.
- Verna and Marrs McLean Department of Biochemistry and Molecular Biology, Baylor College of Medicine, Houston, TX 77030, USA.
Organizational Affiliation: