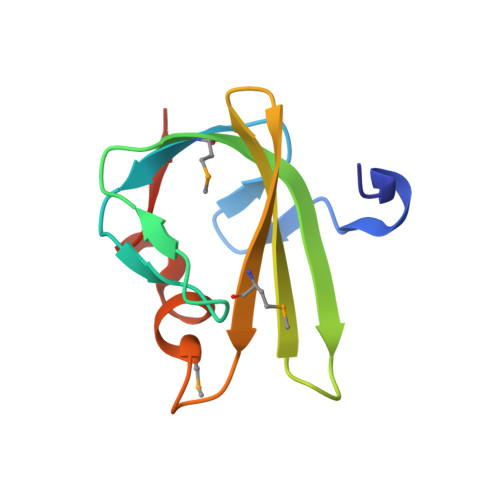
Structure from the mobile metagenome of Vibrio cholerae. Integron cassette protein VCH_CASS4.
Deshpande, C.N., Sureshan, V., Harrop, S.J., Boucher, Y., Xu, X., Cui, H., Edwards, A., Savchenko, A., Joachimiak, A., Tan, K., Stokes, H.W., Curmi, P.M.G., Mabbutt, B.C.To be published.