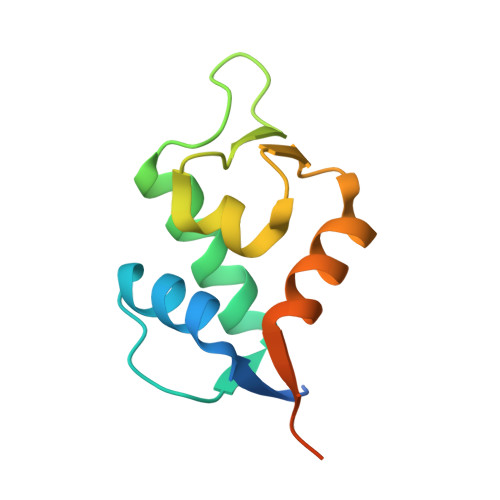
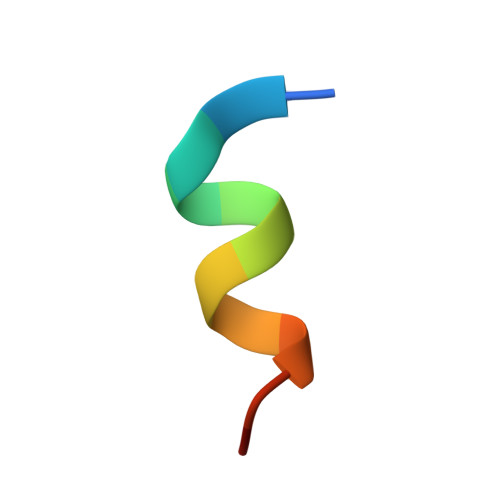High affinity interaction of the p53 peptide-analogue with human Mdm2 and Mdmx.
Czarna, A., Popowicz, G.M., Pecak, A., Wolf, S., Dubin, G., Holak, T.A.(2009) Cell Cycle 8: 1176-1184
- PubMed: 19305137
- DOI: https://doi.org/10.4161/cc.8.8.8185
- Primary Citation of Related Structures:
3FDO, 3G03 - PubMed Abstract:
The Mdm2 and Mdmx proteins are the principal negative regulators of the p53 tumor suppressor. Reactivation of p53 activity by disrupting the Mdm2/Mdmx-p53 interactions offers new possibilities for anticancer therapeutics. Here, we present crystal structures of two complexes, a p53-like mutant peptide with the N-terminal domains of Mdm2 and Mdmx, respectively. The structures reveal that the p53 mutant peptide (amino acid sequence: LTFEHYWAQLTS) assumes virtually identical conformations in both complexes despite the different shapes of the p53-binding pockets in these two proteins, has a more extended helical nature compared to the Mdm2-bound wild-type p53 peptide, and does not disturb the native folds of Mdm2 or Mdmx. The extension of the helical structure in the mutant p53 peptide greatly improves its binding to Mdm2 and Mdmx. The fluorescence polarization assay that we have developed using this peptide indicates the affinities towards Mdm2 of 3.6 nM and for Mdmx of 6.1 nM, compared to the low micromolar binding of a similar length wild-type p53 peptide to Mdm2/Mdmx. Our assay does not require expensive non-native amino acids, and allows measurements of the interaction with both Mdm2 and Mdmx in identical conditions-without modification of experimental conditions or setups between the two proteins. The structural information presented here, coupled with the robust fluorescence polarization assay, should enable development of a simple pharmacophore model of cross-selective Mdm2-Mdmx/p53 inhibitors.
- Max Planck Institute for Biochemistry, Martinsried, Germany.
Organizational Affiliation: