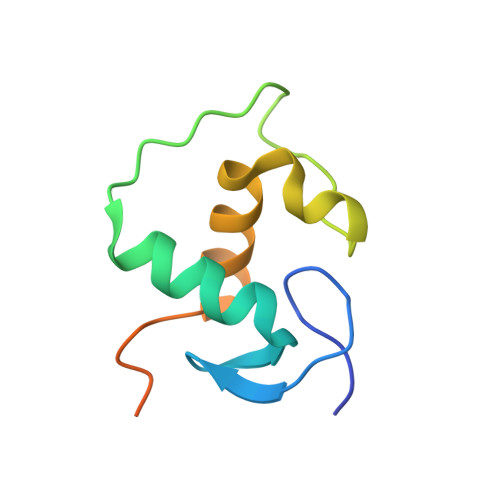
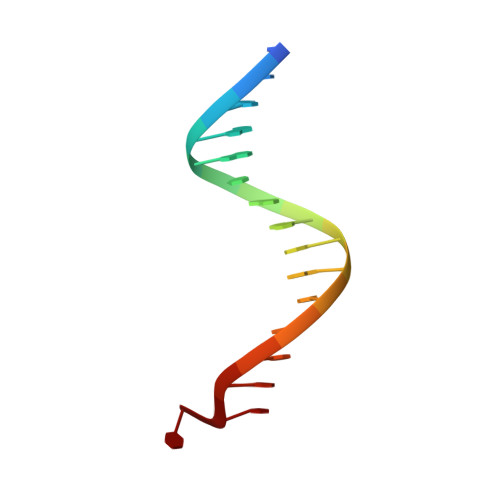
DNA binding site sequence directs glucocorticoid receptor structure and activity.
Meijsing, S.H., Pufall, M.A., So, A.Y., Bates, D.L., Chen, L., Yamamoto, K.R.(2009) Science 324: 407-410
- PubMed: 19372434
- DOI: https://doi.org/10.1126/science.1164265
- Primary Citation of Related Structures:
3FYL, 3G6P, 3G6Q, 3G6R, 3G6T, 3G6U, 3G8U, 3G8X, 3G97, 3G99, 3G9I, 3G9J, 3G9M, 3G9O, 3G9P - PubMed Abstract:
Genes are not simply turned on or off, but instead their expression is fine-tuned to meet the needs of a cell. How genes are modulated so precisely is not well understood. The glucocorticoid receptor (GR) regulates target genes by associating with specific DNA binding sites, the sequences of which differ between genes. Traditionally, these binding sites have been viewed only as docking sites. Using structural, biochemical, and cell-based assays, we show that GR binding sequences, differing by as little as a single base pair, differentially affect GR conformation and regulatory activity. We therefore propose that DNA is a sequence-specific allosteric ligand of GR that tailors the activity of the receptor toward specific target genes.
- Department of Cellular and Molecular Pharmacology, University of California, San Francisco, CA 94158, USA.
Organizational Affiliation: