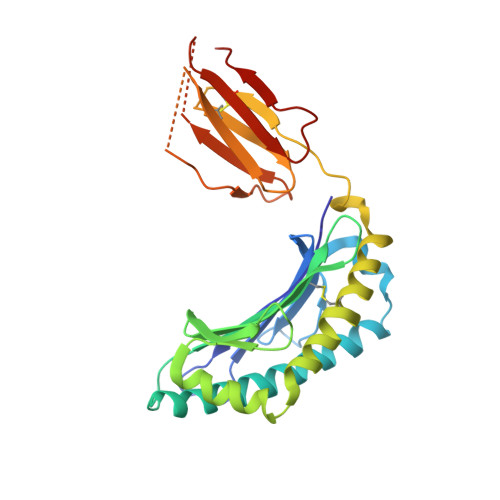
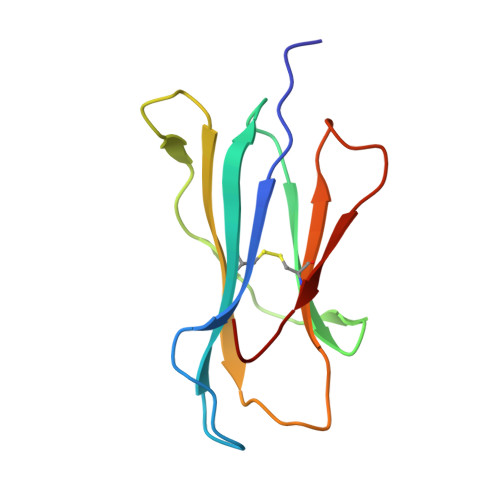
Protective efficacy of cross-reactive CD8+ T cells recognising mutant viral epitopes depends on peptide-MHC-I structural interactions and T cell activation threshold.
Valkenburg, S.A., Gras, S., Guillonneau, C., La Gruta, N.L., Thomas, P.G., Purcell, A.W., Rossjohn, J., Doherty, P.C., Turner, S.J., Kedzierska, K.(2010) PLoS Pathog 6: e1001039-e1001039
- PubMed: 20711359
- DOI: https://doi.org/10.1371/journal.ppat.1001039
- Primary Citation of Related Structures:
3FTG - PubMed Abstract:
Emergence of a new influenza strain leads to a rapid global spread of the virus due to minimal antibody immunity. Pre-existing CD8(+) T-cell immunity directed towards conserved internal viral regions can greatly ameliorate the disease. However, mutational escape within the T cell epitopes is a substantial issue for virus control and vaccine design. Although mutations can result in a loss of T cell recognition, some variants generate cross-reactive T cell responses. In this study, we used reverse genetics to modify the influenza NP(336-374) peptide at a partially-solvent exposed residue (N->A, NPN3A mutation) to assess the availability, effectiveness and mechanism underlying influenza-specific cross-reactive T cell responses. The engineered virus induced a diminished CD8(+) T cell response and selected a narrowed T cell receptor (TCR) repertoire within two V beta regions (V beta 8.3 and V beta 9). This can be partially explained by the H-2D(b)NPN3A structure that showed a loss of several contacts between the NPN3A peptide and H-2D(b), including a contact with His155, a position known to play an important role in mediating TCR-pMHC-I interactions. Despite these differences, common cross-reactive TCRs were detected in both the naïve and immune NPN3A-specific TCR repertoires. However, while the NPN3A epitope primes memory T-cells that give an equivalent recall response to the mutant or wild-type (wt) virus, both are markedly lower than wt->wt challenge. Such decreased CD8(+) responses elicited after heterologous challenge resulted in delayed viral clearance from the infected lung. Furthermore, mice first exposed to the wt virus give a poor, low avidity response following secondary infection with the mutant. Thus, the protective efficacy of cross-reactive CD8(+) T cells recognising mutant viral epitopes depend on peptide-MHC-I structural interactions and functional avidity. Our study does not support vaccine strategies that include immunization against commonly selected cross-reactive variants with mutations at partially-solvent exposed residues that have characteristics comparable to NPN3A.
- Department of Microbiology and Immunology, University of Melbourne, Parkville, Australia.
Organizational Affiliation: