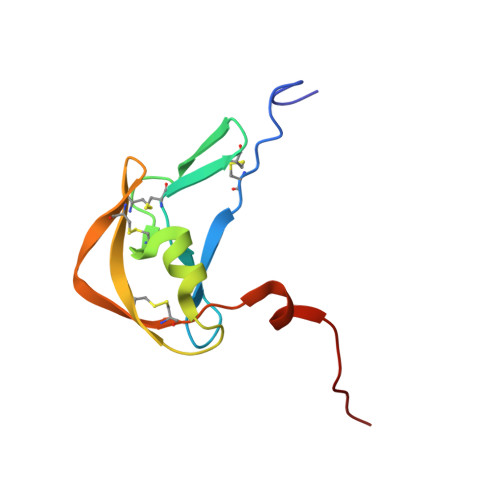
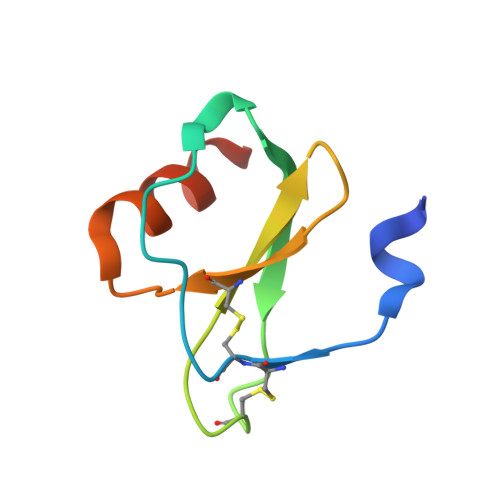
Structural basis of chemokine sequestration by a tick chemokine binding protein: the crystal structure of the complex between Evasin-1 and CCL3
Dias, J.M., Losberger, C., Deruaz, M., Power, C.A., Proudfoot, A.E.I., Shaw, J.P.(2009) PLoS One 4
- PubMed: 20041127
- DOI: https://doi.org/10.1371/journal.pone.0008514
- Primary Citation of Related Structures:
3FPR, 3FPT, 3FPU - PubMed Abstract:
Chemokines are a subset of cytokines responsible for controlling the cellular migration of inflammatory cells through interaction with seven transmembrane G protein-coupled receptors. The blocking of a chemokine-receptor interaction results in a reduced inflammatory response, and represents a possible anti-inflammatory strategy, a strategy that is already employed by some virus and parasites. Anti-chemokine activity has been described in the extracts of tick salivary glands, and we have recently described the cloning and characterization of such chemokine binding proteins from the salivary glands, which we have named Evasins. We have solved the structure of Evasin-1, a very small and highly selective chemokine-binding protein, by x-ray crystallography and report that the structure is novel, with no obvious similarity to the previously described structures of viral chemokine binding proteins. Moreover it does not possess a known fold. We have also solved the structure of the complex of Evasin-1 and its high affinity ligand, CCL3. The complex is a 1:1 heterodimer in which the N-terminal region of CCL3 forms numerous contacts with Evasin-1, including prominent pi-pi interactions between residues Trp89 and Phe14 of the binding protein and Phe29 and Phe13 of the chemokine. However, these interactions do not appear to be crucial for the selectivity of the binding protein, since these residues are found in CCL5, which is not a ligand for Evasin-1. The selectivity of the interaction would appear to lie in the N-terminal residues of the chemokine, which form the "address" whereas the hydrophobic interactions in the rest of the complex would serve primarily to stabilize the complex. A thorough understanding of the binding mode of this small protein, and its other family members, could be very informative in the design of potent neutralizing molecules of pro-inflammatory mediators of the immune system, such as chemokines.
- Merck Serono Geneva Research Center, Merck Serono S.A., Geneva, Switzerland.
Organizational Affiliation: