A Structural Model for the Damage-sensing Complex in Bacterial Nucleotide Excision Repair.
Pakotiprapha, D., Liu, Y., Verdine, G.L., Jeruzalmi, D.(2009) J Biological Chem 284: 12837-12844
- PubMed: 19287003
- DOI: https://doi.org/10.1074/jbc.M900571200
- Primary Citation of Related Structures:
3FPN - PubMed Abstract:
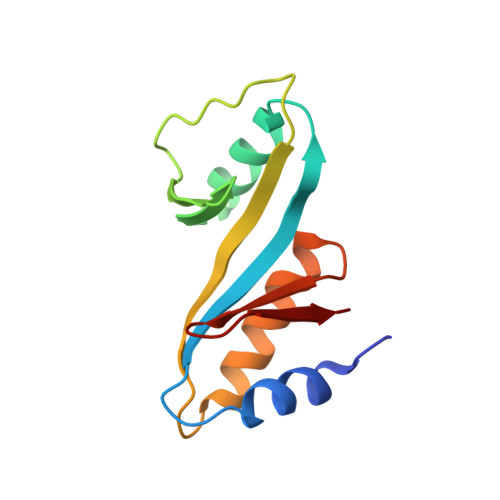
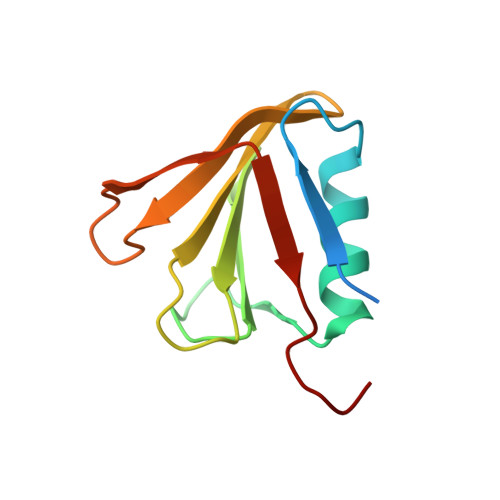
Nucleotide excision repair is distinguished from other DNA repair pathways by its ability to process a wide range of structurally unrelated DNA lesions. In bacteria, damage recognition is achieved by the UvrA.UvrB ensemble. Here, we report the structure of the complex between the interaction domains of UvrA and UvrB. These domains are necessary and sufficient for full-length UvrA and UvrB to associate and thereby form the DNA damage-sensing complex of bacterial nucleotide excision repair. The crystal structure and accompanying biochemical analyses suggest a model for the complete damage-sensing complex.
- Department of Molecular and Cellular Biology, Harvard University, Cambridge, MA 02138, USA.
Organizational Affiliation: