Biochemical and structural properties of chimeras constructed by exchange of cofactor-binding domains in alcohol dehydrogenases from thermophilic and mesophilic microorganisms
Goihberg, E., Peretz, M., Tel-Or, S., Dym, O., Shimon, L., Frolow, F., Burstein, Y.(2010) Biochemistry 49: 1943-1953
- PubMed: 20102159
- DOI: https://doi.org/10.1021/bi901730x
- Primary Citation of Related Structures:
3FPC, 3FPL, 3FSR, 3FTN - PubMed Abstract:
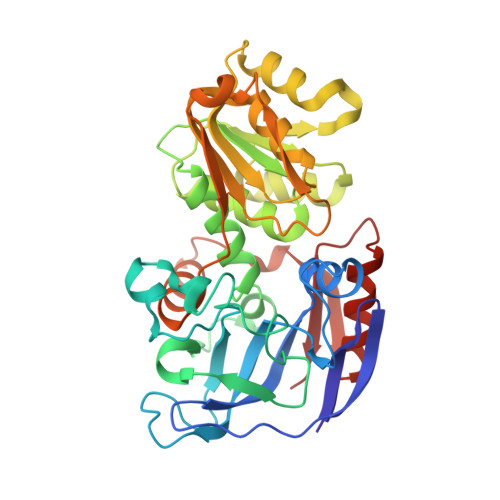
The cofactor-binding domains (residues 153-295) of the alcohol dehydrogenases from the thermophile Thermoanaerobacter brockii (TbADH), the mesophilic bacterium Clostridium beijerinckii (CbADH), and the protozoan parasite Entamoeba histolytica (EhADH1) have been exchanged. Three chimeras have been constructed. In the first chimera, the cofactor-binding domain of thermophilic TbADH was replaced with the cofactor-binding domain of its mesophilic counterpart CbADH [chimera Chi21((TCT))]. This domain exchange significantly destabilized the parent thermophilic enzyme (DeltaT(1/2) = -18 degrees C). The reverse exchange in CbADH [chimera Chi22((CTC))], however, had little effect on the thermal stability of the parent mesophilic protein. Furthermore, substituting the cofactor-binding domain of TbADH with the homologous domain of EhADH1 [chimera Chi23((TET))] substantially reduced the thermal stability of the thermophilic ADH (DeltaT(1/2) = -51 degrees C) and impeded the oligomerization of the enzyme. All three chimeric proteins and one of their site-directed mutants were crystallized, and their three-dimensional (3D) structures were determined. Comparison of the 3D structures of the chimeras and the chimeric mutant with the structures of their parent ADHs showed no significant changes to their Calpha chains, suggesting that the difference in the thermal stability of the three parent ADHs and their chimeric mutants could be due to a limited number of substitutions located at strategic positions, mainly at the oligomerization interfaces. Indeed, stabilization of the chimeras was achieved, to a significant extent, either by introduction of a proline residue at a strategic position in the major horse liver ADH-type dimerization interface (DeltaT(1/2) = 35 degrees C) or by introduction of intersubunit electrostatic interactions (DeltaT(1/2) = 6 degrees C).
- Department of Organic Chemistry, Weizmann Institute of Science, Rehovot 76100, Israel.
Organizational Affiliation: