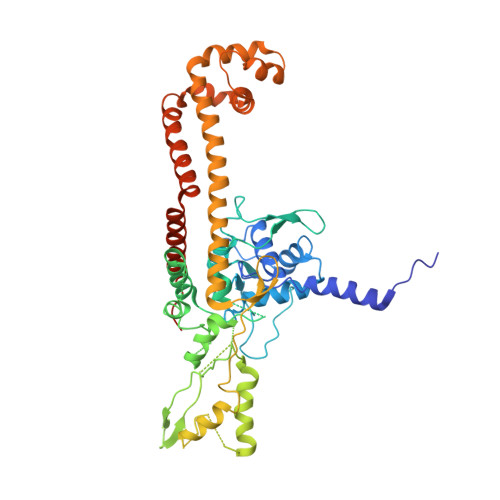
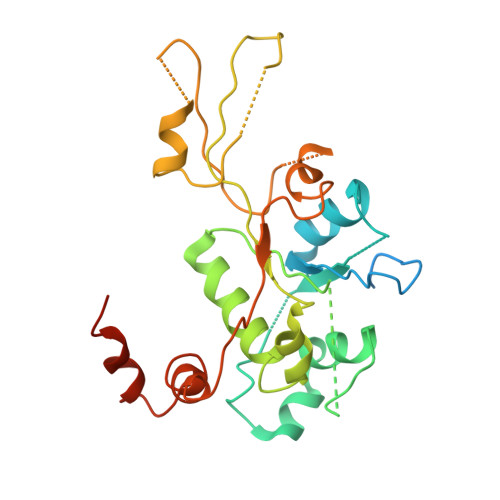
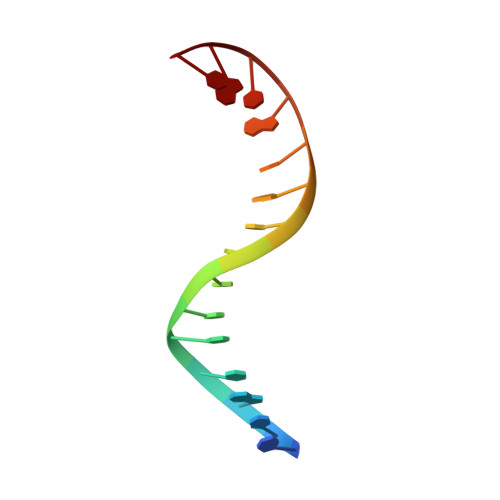
Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases
Laponogov, I., Sohi, M.K., Veselkov, D.A., Pan, X.-S., Sawhney, R., Thompson, A.W., McAuley, K.E., Fisher, L.M., Sanderson, M.R.(2009) Nat Struct Mol Biol 16: 667-669
- PubMed: 19448616
- DOI: https://doi.org/10.1038/nsmb.1604
- Primary Citation of Related Structures:
3FOE, 3FOF - PubMed Abstract:
Type II topoisomerases alter DNA topology by forming a covalent DNA-cleavage complex that allows DNA transport through a double-stranded DNA break. We present the structures of cleavage complexes formed by the Streptococcus pneumoniae ParC breakage-reunion and ParE TOPRIM domains of topoisomerase IV stabilized by moxifloxacin and clinafloxacin, two antipneumococcal fluoroquinolones. These structures reveal two drug molecules intercalated at the highly bent DNA gate and help explain antibacterial quinolone action and resistance.
- Randall Division of Cell and Molecular Biophysics, King's College London, University of London, London, UK.
Organizational Affiliation: