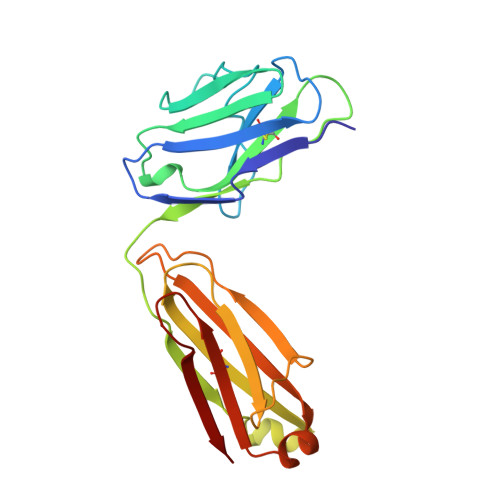
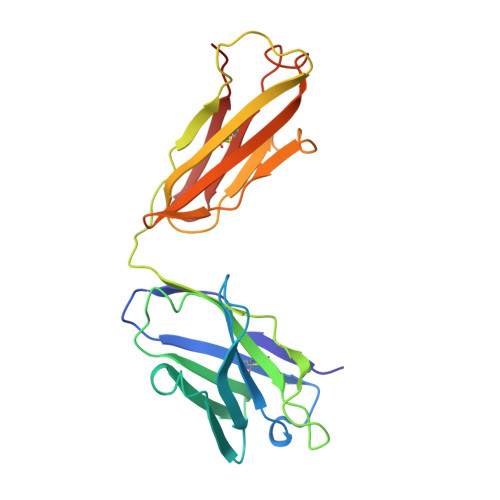
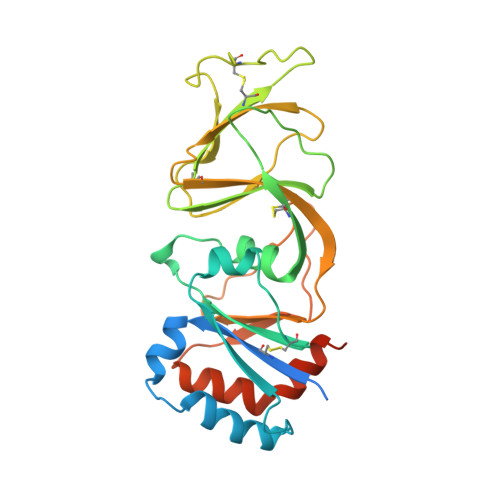
Structure of rotavirus outer-layer protein VP7 bound with a neutralizing Fab.
Aoki, S.T., Settembre, E.C., Trask, S.D., Greenberg, H.B., Harrison, S.C., Dormitzer, P.R.(2009) Science 324: 1444-1447
- PubMed: 19520960
- DOI: https://doi.org/10.1126/science.1170481
- Primary Citation of Related Structures:
3FMG - PubMed Abstract:
Rotavirus outer-layer protein VP7 is a principal target of protective antibodies. Removal of free calcium ions (Ca2+) dissociates VP7 trimers into monomers, releasing VP7 from the virion, and initiates penetration-inducing conformational changes in the other outer-layer protein, VP4. We report the crystal structure at 3.4 angstrom resolution of VP7 bound with the Fab fragment of a neutralizing monoclonal antibody. The Fab binds across the outer surface of the intersubunit contact, which contains two Ca2+ sites. Mutations that escape neutralization by other antibodies suggest that the same region bears the epitopes of most neutralizing antibodies. The monovalent Fab is sufficient to neutralize infectivity. We propose that neutralizing antibodies against VP7 act by stabilizing the trimer, thereby inhibiting the uncoating trigger for VP4 rearrangement. A disulfide-linked trimer is a potential subunit immunogen.
- Laboratory of Molecular Medicine, Children's Hospital, Boston, MA 02115, USA.
Organizational Affiliation: